The Exploration of Novel Regulatory Relationships Drives Haloarchaeal Operon-Like Structural Dynamics over Short Evolutionary Distances
- PMID: 33266086
- PMCID: PMC7760734
- DOI: 10.3390/microorganisms8121900
The Exploration of Novel Regulatory Relationships Drives Haloarchaeal Operon-Like Structural Dynamics over Short Evolutionary Distances
Abstract
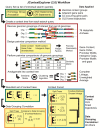
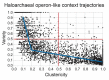
Operons are a dominant feature of bacterial and archaeal genome organization. Numerous investigations have related aspects of operon structure to operon function, making operons exemplars for studies aimed at deciphering Nature's design principles for genomic organization at a local scale. We consider this understanding to be both fundamentally important and ultimately useful in the de novo design of increasingly complex synthetic circuits. Here we analyze the evolution of the genomic context of operon-like structures in a set of 76 sequenced and annotated species of halophilic archaea. The phylogenetic depth and breadth of this dataset allows insight into changes in operon-like structures over shorter evolutionary time scales than have been studied in previous cross-species analysis of operon evolution. Our analysis, implemented in the updated software package JContextExplorer finds that operon-like context as measured by changes in structure frequently differs from a sequence divergence model of whole-species phylogeny and that changes seem to be dominated by the exploration of novel regulatory relationships.
Keywords: archaea; evolution; genomics; haloarchaea; operon.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures







References
-
- Hillier L.W., Miller R.D., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. - DOI - PMC - PubMed

