Broad and Complex Roles of NBR1-Mediated Selective Autophagy in Plant Stress Responses
- PMID: 33266087
- PMCID: PMC7760648
- DOI: 10.3390/cells9122562
Broad and Complex Roles of NBR1-Mediated Selective Autophagy in Plant Stress Responses
Abstract
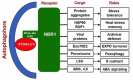
Selective autophagy is a highly regulated degradation pathway for the removal of specific damaged or unwanted cellular components and organelles such as protein aggregates. Cargo selectivity in selective autophagy relies on the action of cargo receptors and adaptors. In mammalian cells, two structurally related proteins p62 and NBR1 act as cargo receptors for selective autophagy of ubiquitinated proteins including aggregation-prone proteins in aggrephagy. Plant NBR1 is the structural and functional homolog of mammalian p62 and NBR1. Since its first reports almost ten years ago, plant NBR1 has been well established to function as a cargo receptor for selective autophagy of stress-induced protein aggregates and play an important role in plant responses to a broad spectrum of stress conditions including heat, salt and drought. Over the past several years, important progress has been made in the discovery of specific cargo proteins of plant NBR1 and their roles in the regulation of plant heat stress memory, plant-viral interaction and special protein secretion. There is also new evidence for a possible role of NBR1 in stress-induced pexophagy, sulfur nutrient responses and abscisic acid signaling. In this review, we summarize these progresses and discuss the potential significance of NBR1-mediated selective autophagy in broad plant responses to both biotic and abiotic stresses.
Keywords: NBR1; autophagy; plant heat tolerance; plant stress responses; plant virus interaction; protein aggregates; selective autophagy receptor.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
NBR1: The archetypal selective autophagy receptor.J Cell Biol. 2022 Nov 7;221(11):e202208092. doi: 10.1083/jcb.202208092. Epub 2022 Oct 18. J Cell Biol. 2022. PMID: 36255390 Free PMC article. Review.
-
NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses.PLoS Genet. 2013;9(1):e1003196. doi: 10.1371/journal.pgen.1003196. Epub 2013 Jan 17. PLoS Genet. 2013. PMID: 23341779 Free PMC article.
-
Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins.J Exp Bot. 2020 Jan 1;71(1):73-89. doi: 10.1093/jxb/erz404. J Exp Bot. 2020. PMID: 31494674 Free PMC article.
-
Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1.Autophagy. 2011 Sep;7(9):993-1010. doi: 10.4161/auto.7.9.16389. Epub 2011 Sep 1. Autophagy. 2011. PMID: 21606687 Free PMC article.
-
Cargo Recognition and Function of Selective Autophagy Receptors in Plants.Int J Mol Sci. 2021 Jan 20;22(3):1013. doi: 10.3390/ijms22031013. Int J Mol Sci. 2021. PMID: 33498336 Free PMC article. Review.
Cited by
-
Editorial: Highlights in Autophagy-From Basic Mechanisms to Human Disorder Treatments.Cells. 2023 Jan 3;12(1):188. doi: 10.3390/cells12010188. Cells. 2023. PMID: 36611981 Free PMC article.
-
The Valsa Mali effector Vm1G-1794 protects the aggregated MdEF-Tu from autophagic degradation to promote infection in apple.Autophagy. 2023 Jun;19(6):1745-1763. doi: 10.1080/15548627.2022.2153573. Epub 2022 Dec 7. Autophagy. 2023. PMID: 36449354 Free PMC article.
-
NBR1: The archetypal selective autophagy receptor.J Cell Biol. 2022 Nov 7;221(11):e202208092. doi: 10.1083/jcb.202208092. Epub 2022 Oct 18. J Cell Biol. 2022. PMID: 36255390 Free PMC article. Review.
-
ATG8-Interacting Motif: Evolution and Function in Selective Autophagy of Targeting Biological Processes.Front Plant Sci. 2021 Nov 29;12:783881. doi: 10.3389/fpls.2021.783881. eCollection 2021. Front Plant Sci. 2021. PMID: 34912364 Free PMC article. Review.
-
Subversion of selective autophagy for the biogenesis of tombusvirus replication organelles inhibits autophagy.PLoS Pathog. 2024 Mar 14;20(3):e1012085. doi: 10.1371/journal.ppat.1012085. eCollection 2024 Mar. PLoS Pathog. 2024. PMID: 38484009 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

