Bioprospecting for Novel Halophilic and Halotolerant Sources of Hydrolytic Enzymes in Brackish, Saline and Hypersaline Lakes of Romania
- PMID: 33266166
- PMCID: PMC7760675
- DOI: 10.3390/microorganisms8121903
Bioprospecting for Novel Halophilic and Halotolerant Sources of Hydrolytic Enzymes in Brackish, Saline and Hypersaline Lakes of Romania
Abstract
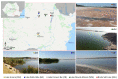
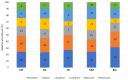
Halophilic and halotolerant microorganisms represent promising sources of salt-tolerant enzymes that could be used in various biotechnological processes where high salt concentrations would otherwise inhibit enzymatic transformations. Considering the current need for more efficient biocatalysts, the present study aimed to explore the microbial diversity of five under- or uninvestigated salty lakes in Romania for novel sources of hydrolytic enzymes. Bacteria, archaea and fungi were obtained by culture-based approaches and screened for the production of six hydrolases (protease, lipase, amylase, cellulase, xylanase and pectinase) using agar plate-based assays. Moreover, the phylogeny of bacterial and archaeal isolates was studied through molecular methods. From a total of 244 microbial isolates, 182 (74.6%) were represented by bacteria, 22 (9%) by archaea, and 40 (16.4%) by fungi. While most bacteria synthesized protease and lipase, the most frequent hydrolase produced by fungi was pectinase. The archaeal isolates had limited hydrolytic activity, being able to produce only amylase and cellulase. Among the taxonomically identified isolates, the best hydrolytic activities were observed in halotolerant bacteria belonging to the genus Bacillus and in extremely halophilic archaea of the genera Haloterrigena and Halostagnicola. Therefore, the present study highlights that the investigated lakes harbor various promising species of microorganisms able to produce industrially valuable enzymes.
Keywords: extracellular hydrolases; extreme environments; extremozymes; halophiles; halophilic archaea; halotolerant bacteria; hydrolytic enzymes; hypersaline lakes; salt-tolerant enzymes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- McGenity T.J., Oren A. Hypersaline Environments. In: Bell E.M., editor. Life at Extremes: Environments, Organisms and Strategies for Survival. CAB International; Oxfordshire, UK: 2012. pp. 402–437. - DOI
-
- Oren A. Life in Hypersaline Environments. In: Hurst C.J., editor. Their World: A Diversity of Microbial Environments, Advances in Environmental Microbiology. Springer International Publishing; Cham, Switzerland: 2016. pp. 301–339. - DOI
-
- Ventosa A., Márquez M.C., Sánchez-Porro C., de la Haba R.R. Taxonomy of Halophilic Archaea and Bacteria. In: Vreeland R.H., editor. Advances in Understanding the Biology of Halophilic Microorganisms. Springer; Dordrecht, The Netherlands: 2012. pp. 59–80. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

