Mechanistic Insights into the Allosteric Regulation of the Clr4 Protein Lysine Methyltransferase by Autoinhibition and Automethylation
- PMID: 33266419
- PMCID: PMC7700585
- DOI: 10.3390/ijms21228832
Mechanistic Insights into the Allosteric Regulation of the Clr4 Protein Lysine Methyltransferase by Autoinhibition and Automethylation
Abstract
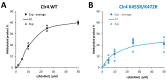
Clr4 is a histone H3 lysine 9 methyltransferase in Schizosaccharomyces pombe that is essential for heterochromatin formation. Previous biochemical and structural studies have shown that Clr4 is in an autoinhibited state in which an autoregulatory loop (ARL) blocks the active site. Automethylation of lysine residues in the ARL relieves autoinhibition. To investigate the mechanism of Clr4 regulation by autoinhibition and automethylation, we exchanged residues in the ARL by site-directed mutagenesis leading to stimulation or inhibition of automethylation and corresponding changes in Clr4 catalytic activity. Furthermore, we demonstrate that Clr4 prefers monomethylated (H3K9me1) over unmodified (H3K9me0) histone peptide substrates, similar to related human enzymes and, accordingly, H3K9me1 is more efficient in overcoming autoinhibition. Due to enzyme activation by automethylation, we observed a sigmoidal dependence of Clr4 activity on the AdoMet concentration, with stimulation at high AdoMet levels. In contrast, an automethylation-deficient mutant showed a hyperbolic Michaelis-Menten type relationship. These data suggest that automethylation of the ARL could act as a sensor for AdoMet levels in cells and regulate the generation and maintenance of heterochromatin accordingly. This process could connect epigenome modifications with the metabolic state of cells. As other human protein lysine methyltransferases (for example, PRC2) also use automethylation/autoinhibition mechanisms, our results may provide a model to describe their regulation as well.
Keywords: automethylation; enzyme kinetics; enzyme regulation; protein methylation; protein methyltransferase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

