Machine Learning Identifies Robust Matrisome Markers and Regulatory Mechanisms in Cancer
- PMID: 33266472
- PMCID: PMC7700160
- DOI: 10.3390/ijms21228837
Machine Learning Identifies Robust Matrisome Markers and Regulatory Mechanisms in Cancer
Abstract
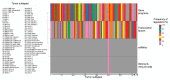
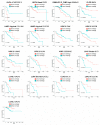
The expression and regulation of matrisome genes-the ensemble of extracellular matrix, ECM, ECM-associated proteins and regulators as well as cytokines, chemokines and growth factors-is of paramount importance for many biological processes and signals within the tumor microenvironment. The availability of large and diverse multi-omics data enables mapping and understanding of the regulatory circuitry governing the tumor matrisome to an unprecedented level, though such a volume of information requires robust approaches to data analysis and integration. In this study, we show that combining Pan-Cancer expression data from The Cancer Genome Atlas (TCGA) with genomics, epigenomics and microenvironmental features from TCGA and other sources enables the identification of "landmark" matrisome genes and machine learning-based reconstruction of their regulatory networks in 74 clinical and molecular subtypes of human cancers and approx. 6700 patients. These results, enriched for prognostic genes and cross-validated markers at the protein level, unravel the role of genetic and epigenetic programs in governing the tumor matrisome and allow the prioritization of tumor-specific matrisome genes (and their regulators) for the development of novel therapeutic approaches.
Keywords: big data; bioinformatics; cancer; extracellular matrix; matrisome; regulatory networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rianna C., Kumar P., Radmacher M. The Role of the Microenvironment in the Biophysics of Cancer. Semin. Cell Dev. Biol. 2018;73:107–114. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

