Detection of CRISPR-mediated genome modifications through altered methylation patterns of CpG islands
- PMID: 33267773
- PMCID: PMC7709351
- DOI: 10.1186/s12864-020-07233-2
Detection of CRISPR-mediated genome modifications through altered methylation patterns of CpG islands
Abstract
Background: The development and application of CRISPR technologies for the modification of the genome are rapidly expanding. Advances in the field describe new CRISPR components that are strategically engineered to improve the precision and reliability of CRISPR editing within the genome sequence. Genome modification using induced genome breaks that are targeted and mediated by CRISPR components leverage cellular mechanisms for repair like homology directed repair (HDR) to incorporate genomic edits with increased precision.
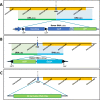
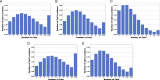
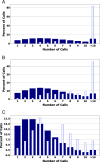
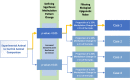
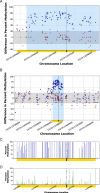
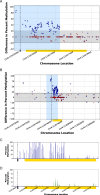
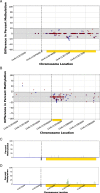
Results: In this report, we describe the gain of methylation at typically hypomethylated CpG island (CGI) locations affected by the CRISPR-mediated incorporation of donor DNA using HDR mechanisms. With characterization of CpG methylation patterns using whole genome bisulfite sequencing, these CGI methylation disruptions trace the insertion of the donor DNA during the genomic edit. These insertions mediated by homology-directed recombination disrupt the generational methylation pattern stability of the edited CGI within the cells and their cellular lineage within the animal strain, persisting across generations. Our approach describes a statistically based workflow for indicating locations of modified CGIs and provides a mechanism for evaluating the directed modification of the methylome of the affected CGI at the CpG-level.
Conclusions: With advances in genome modification technology comes the need to detect the level and persistence of methylation change that modifications to the genomic sequence impose upon the collaterally edited methylome. Any modification of the methylome of somatic or germline cells could have implications for gene regulation mechanisms governed by the methylation patterns of CGI regions in the application of therapeutic edits of more sensitively regulated genomic regions. The method described here locates the directed modification of the mouse epigenome that persists over generations. While this observance would require supporting molecular observations such as direct sequence changes or gene expression changes, the observation of epigenetic modification provides an indicator that intentionally directed genomic edits can lead to collateral, unintentional epigenomic changes post modification with generational persistence.
Keywords: CRISPR genome editing; CpG island; Epigenetic modification; Homology-directed repair; Methylation variance; Non-homologous end-joining; Statistical variance detection.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

