Transcriptomic profiles of non-embryogenic and embryogenic callus cells in a highly regenerative upland cotton line (Gossypium hirsutum L.)
- PMID: 33267776
- PMCID: PMC7713314
- DOI: 10.1186/s12861-020-00230-4
Transcriptomic profiles of non-embryogenic and embryogenic callus cells in a highly regenerative upland cotton line (Gossypium hirsutum L.)
Abstract
Background: Genotype independent transformation and whole plant regeneration through somatic embryogenesis relies heavily on the intrinsic ability of a genotype to regenerate. The critical genetic architecture of non-embryogenic callus (NEC) cells and embryogenic callus (EC) cells in a highly regenerable cotton genotype is unknown.
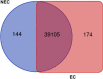
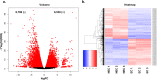
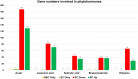
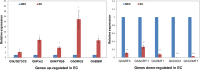
Results: In this study, gene expression profiles of a highly regenerable Gossypium hirsutum L. cultivar, Jin668, were analyzed at two critical developmental stages during somatic embryogenesis, non-embryogenic callus (NEC) cells and embryogenic callus (EC) cells. The rate of EC formation in Jin668 is 96%. Differential gene expression analysis revealed a total of 5333 differentially expressed genes (DEG) with 2534 genes upregulated and 2799 genes downregulated in EC. A total of 144 genes were unique to NEC cells and 174 genes were unique to EC. Clustering and enrichment analysis identified genes upregulated in EC that function as transcription factors/DNA binding, phytohormone response, oxidative reduction, and regulators of transcription; while genes categorized in methylation pathways were downregulated. Four key transcription factors were identified based on their sharp upregulation in EC tissue; LEAFY COTYLEDON 1 (LEC1), BABY BOOM (BBM), FUSCA (FUS3) and AGAMOUS-LIKE15 with distinguishable subgenome expression bias.
Conclusions: This comparative analysis of NEC and EC transcriptomes gives new insights into the genes involved in somatic embryogenesis in cotton.
Keywords: Callus, embryo; Gossypium hirsutum L; Somatic embryogenesis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Dunwell JM. Transgenic approaches to crop improvement. J Exp Bot 2000; 51 Spec No:487–496. - PubMed
-
- Mccabe DE, Swain WF, Martinell BJ, Christou P. Stable transformation of soybean (glycine-max) by particle-acceleration. Bio-Technol. 1988;6(8):923–926.
-
- Cho MJ, Yano H, Okamoto D, Kim HK, Jung HR, Newcomb K, Le VK, Yoo HS, Langham R, Buchanan BB, Lemaux PG. Stable transformation of rice (Oryza sativa L.) via microprojectile bombardment of highly regenerative, green tissues derived from mature seed. Plant Cell Rep. 2004;22(7):483–489. doi: 10.1007/s00299-003-0713-7. - DOI - PubMed
-
- Gordon-Kamm WJ, Spencer TM, Mangano ML, Adams TR, Daines RJ, Start WG, O'Brien JV, Chambers SA, Adams WR, Jr, Willetts NG, Rice TB, Mackey CJ, Krueger RW, Kausch AP, Lemaux PG. Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell. 1990;2(7):603–618. doi: 10.2307/3869124. - DOI - PMC - PubMed
-
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, Lemaux PG, Medford JI, Orozco-Cardenas ML, Tricoli DM, Van Eck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart CN., Jr Advancing crop transformation in the era of genome editing. Plant Cell. 2016;28(7):1510–1520. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

