Histone H3.3 beyond cancer: Germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients
- PMID: 33268356
- PMCID: PMC7821880
- DOI: 10.1126/sciadv.abc9207
Histone H3.3 beyond cancer: Germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients
Abstract
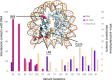
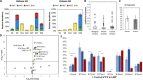
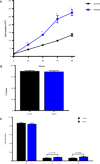
Although somatic mutations in Histone 3.3 (H3.3) are well-studied drivers of oncogenesis, the role of germline mutations remains unreported. We analyze 46 patients bearing de novo germline mutations in histone 3 family 3A (H3F3A) or H3F3B with progressive neurologic dysfunction and congenital anomalies without malignancies. Molecular modeling of all 37 variants demonstrated clear disruptions in interactions with DNA, other histones, and histone chaperone proteins. Patient histone posttranslational modifications (PTMs) analysis revealed notably aberrant local PTM patterns distinct from the somatic lysine mutations that cause global PTM dysregulation. RNA sequencing on patient cells demonstrated up-regulated gene expression related to mitosis and cell division, and cellular assays confirmed an increased proliferative capacity. A zebrafish model showed craniofacial anomalies and a defect in Foxd3-derived glia. These data suggest that the mechanism of germline mutations are distinct from cancer-associated somatic histone mutations but may converge on control of cell proliferation.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Bagchi R. A., Weeks K. L., Histone deacetylases in cardiovascular and metabolic diseases. J. Mol. Cell. Cardiol. 130, 151–159 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

