Resurrecting the ancient glow of the fireflies
- PMID: 33268373
- PMCID: PMC7710365
- DOI: 10.1126/sciadv.abc5705
Resurrecting the ancient glow of the fireflies
Abstract
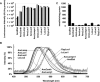
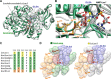
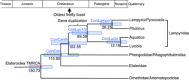
The color of firefly bioluminescence is determined by the structure of luciferase. Firefly luciferase genes have been isolated from more than 30 species, producing light ranging in color from green to orange-yellow. Here, we reconstructed seven ancestral firefly luciferase genes, characterized the enzymatic properties of the recombinant proteins, and determined the crystal structures of the gene from ancestral Lampyridae. Results showed that the synthetic luciferase for the last common firefly ancestor exhibited green light caused by a spatial constraint on the luciferin molecule in enzyme, while fatty acyl-CoA synthetic activity, an original function of firefly luciferase, was diminished in exchange. All known firefly species are bioluminescent in the larvae, with a common ancestor arising approximately 100 million years ago. Combined, our findings propose that, within the mid-Cretaceous forest, the common ancestor of fireflies evolved green light luciferase via trade-off of the original function, which was likely aposematic warning display against nocturnal predation.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- S. Lewis, Silent Sparks: The Wondrous World of Fireflies (Princeton Univ. Press, 2016).
-
- Martin G. J., Branham M. A., Whiting M. F., Bybee S. M., Total evidence phylogeny and the evolution of adult bioluminescence in fireflies (Coleoptera: Lampyridae). Mol. Phylogenet. Evol. 107, 564–575 (2017). - PubMed
-
- Martin G. J., Stanger-Hall K. F., Branham M. A., Da Silveira L. F. L., Lower S. E., Hall D. W., Li X.-Y., Lemmon A. R., Lemmon E. M., Bybee S. M., Higher-level phylogeny and reclassification of Lampyridae (Coleoptera: Elateroidea). Insect Syst. Diver. 3, 11 (2019).
-
- Fallon T. R., Lower S. E., Chang C.-H., Bessho-Uehara M., Martin G. J., Bewick A. J., Behringer M., Debat H. J., Wong I., Day J. C., Suvorov A., Silva C. J., Stanger-Hall K. F., Hall D. W., Schmitz R. J., Nelson D. R., Lewis S. M., Shigenobu S., Bybee S. M., Larracuente A. M., Oba Y., Weng J.-K., Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife 7, e36495 (2018). - PMC - PubMed
-
- Seliger H. H., Lall A. B., Lloyd J. E., Biggley W. H., The colors of firefly bioluminescence—I. Optimization model. Photochem. Photobiol. 36, 673–680 (1982).
Publication types
LinkOut - more resources
Full Text Sources

