EPySeg: a coding-free solution for automated segmentation of epithelia using deep learning
- PMID: 33268451
- PMCID: PMC7774881
- DOI: 10.1242/dev.194589
EPySeg: a coding-free solution for automated segmentation of epithelia using deep learning
Abstract
Epithelia are dynamic tissues that self-remodel during their development. During morphogenesis, the tissue-scale organization of epithelia is obtained through a sum of individual contributions of the cells constituting the tissue. Therefore, understanding any morphogenetic event first requires a thorough segmentation of its constituent cells. This task, however, usually involves extensive manual correction, even with semi-automated tools. Here, we present EPySeg, an open-source, coding-free software that uses deep learning to segment membrane-stained epithelial tissues automatically and very efficiently. EPySeg, which comes with a straightforward graphical user interface, can be used as a Python package on a local computer, or on the cloud via Google Colab for users not equipped with deep-learning compatible hardware. By substantially reducing human input in image segmentation, EPySeg accelerates and improves the characterization of epithelial tissues for all developmental biologists.
Keywords: Computer vision; Deep learning; Epithelia; Quantitative biology; Segmentation; Software.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
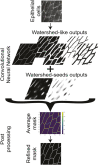
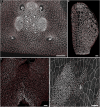
References
-
- Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., Corrado G. S., Davis A., Dean J., Devin M. et al. (2016). TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint, arXiv:1603.04467 [cs.DC].
-
- Aigouy B., Umetsu D. and Eaton S. (2016). Segmentation and Quantitative Analysis of Epithelial Tissues. In Drosophila: Methods and Protocols ( Dahmann C.), pp. 227-239. New York: Springer. - PubMed
-
- Buchholz T.-O., Prakash M., Krull A. and Jug F. (2020). DenoiSeg: joint denoising and segmentation. arXiv preprint, arXiv:2005.02987 [cs.CV].
-
- Chaurasia A. and Culurciello E (2017). LinkNet: Exploiting encoder representations for efficient semantic segmentation. In 2017 IEEE Visual Communications and Image Processing (VCIP), pp. 1-4. IEEE; 10.1109/VCIP.2017.8305148 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

