The Fiber Knob Protein of Human Adenovirus Type 49 Mediates Highly Efficient and Promiscuous Infection of Cancer Cell Lines Using a Novel Cell Entry Mechanism
- PMID: 33268514
- PMCID: PMC7851562
- DOI: 10.1128/JVI.01849-20
The Fiber Knob Protein of Human Adenovirus Type 49 Mediates Highly Efficient and Promiscuous Infection of Cancer Cell Lines Using a Novel Cell Entry Mechanism
Abstract
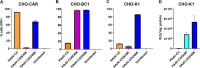
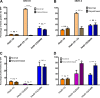
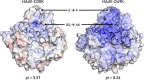
The human adenovirus (HAdV) phylogenetic tree is diverse, divided across seven species and comprising over 100 individual types. Species D HAdV are rarely isolated with low rates of preexisting immunity, making them appealing for therapeutic applications. Several species D vectors have been developed as vaccines against infectious diseases, where they induce robust immunity in preclinical models and early phase clinical trials. However, many aspects of the basic virology of species D HAdV, including their basic receptor usage and means of cell entry, remain understudied. Here, we investigated HAdV-D49, which previously has been studied for vaccine and vascular gene transfer applications. We generated a pseudotyped HAdV-C5 presenting the HAdV-D49 fiber knob protein (HAdV-C5/D49K). This pseudotyped vector was efficient at infecting cells devoid of all known HAdV receptors, indicating HAdV-D49 uses an unidentified cellular receptor. Conversely, a pseudotyped vector presenting the fiber knob protein of the closely related HAdV-D30 (HAdV-C5/D30K), differing in four amino acids from HAdV-D49, failed to demonstrate the same tropism. These four amino acid changes resulted in a change in isoelectric point of the knob protein, with HAdV-D49K possessing a basic apical region compared to a more acidic region in HAdV-D30K. Structurally and biologically we demonstrate that HAdV-D49 knob protein is unable to engage CD46, while potential interaction with coxsackievirus and adenovirus receptor (CAR) is extremely limited by extension of the DG loop. HAdV-C5/49K efficiently transduced cancer cell lines of pancreatic, breast, lung, esophageal, and ovarian origin, indicating it may have potential for oncolytic virotherapy applications, especially for difficult to transduce tumor types.IMPORTANCE Adenoviruses are powerful tools experimentally and clinically. To maximize efficacy, the development of serotypes with low preexisting levels of immunity in the population is desirable. Consequently, attention has focused on those derived from species D, which have proven robust vaccine platforms. This widespread usage is despite limited knowledge in their basic biology and cellular tropism. We investigated the tropism of HAdV-D49, demonstrating that it uses a novel cell entry mechanism that bypasses all known HAdV receptors. We demonstrate, biologically, that a pseudotyped HAdV-C5/D49K vector efficiently transduces a wide range of cell lines, including those presenting no known adenovirus receptor. Structural investigation suggests that this broad tropism is the result of a highly basic electrostatic surface potential, since a homologous pseudotyped vector with a more acidic surface potential, HAdV-C5/D30K, does not display a similar pantropism. Therefore, HAdV-C5/D49K may form a powerful vector for therapeutic applications capable of infecting difficult to transduce cells.
Keywords: adenoviruses; anticancer therapy; oncolytic viruses; surface receptor.
Copyright © 2021 Baker et al.
Figures


Similar articles
-
Pseudotyped αvβ6 integrin-targeted adenovirus vectors for ovarian cancer therapies.Oncotarget. 2016 May 10;7(19):27926-37. doi: 10.18632/oncotarget.8545. Oncotarget. 2016. PMID: 27056886 Free PMC article.
-
Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2020732118. doi: 10.1073/pnas.2020732118. Proc Natl Acad Sci U S A. 2021. PMID: 33384338 Free PMC article.
-
Selection Pressure in the Human Adenovirus Fiber Knob Drives Cell Specificity in Epidemic Keratoconjunctivitis.J Virol. 2016 Oct 14;90(21):9598-9607. doi: 10.1128/JVI.01010-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27512073 Free PMC article.
-
Tropism and transduction of oncolytic adenovirus 5 vectors in cancer therapy: Focus on fiber chimerism and mosaicism, hexon and pIX.Virus Res. 2018 Sep 15;257:40-51. doi: 10.1016/j.virusres.2018.08.012. Epub 2018 Aug 17. Virus Res. 2018. PMID: 30125593 Review.
-
Adenovirus Receptor Expression in Cancer and Its Multifaceted Role in Oncolytic Adenovirus Therapy.Int J Mol Sci. 2020 Sep 17;21(18):6828. doi: 10.3390/ijms21186828. Int J Mol Sci. 2020. PMID: 32957644 Free PMC article. Review.
Cited by
-
Many locks to one key: N-acetylneuraminic acid binding to proteins.IUCrJ. 2024 Sep 1;11(Pt 5):664-674. doi: 10.1107/S2052252524005360. IUCrJ. 2024. PMID: 38965900 Free PMC article. Review.
-
Engineering Adenoviral Vectors with Improved GBM Selectivity.Viruses. 2023 Apr 28;15(5):1086. doi: 10.3390/v15051086. Viruses. 2023. PMID: 37243172 Free PMC article.
-
The Immune System-A Double-Edged Sword for Adenovirus-Based Therapies.Viruses. 2024 Jun 17;16(6):973. doi: 10.3390/v16060973. Viruses. 2024. PMID: 38932265 Free PMC article. Review.
-
A pseudotyped adenovirus serotype 5 vector with serotype 49 fiber knob is an effective vector for vaccine and gene therapy applications.Mol Ther Methods Clin Dev. 2024 Jul 30;32(3):101308. doi: 10.1016/j.omtm.2024.101308. eCollection 2024 Sep 12. Mol Ther Methods Clin Dev. 2024. PMID: 39206304 Free PMC article.
-
Adenovirus Structure: What Is New?Int J Mol Sci. 2021 May 15;22(10):5240. doi: 10.3390/ijms22105240. Int J Mol Sci. 2021. PMID: 34063479 Free PMC article. Review.
References
-
- Alba R, Baker Andrew H, Nicklin Stuart A. 2012. Vector systems for prenatal gene therapy: principles of adenovirus design and production, p 55–84. In Coutelle C, Waddington SN (ed), Prenatal gene therapy. Humana Press, New York, NY. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

