Human Cytomegalovirus Induces the Expression of the AMPKa2 Subunit to Drive Glycolytic Activation and Support Productive Viral Infection
- PMID: 33268515
- PMCID: PMC8092818
- DOI: 10.1128/JVI.01321-20
Human Cytomegalovirus Induces the Expression of the AMPKa2 Subunit to Drive Glycolytic Activation and Support Productive Viral Infection
Abstract
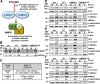
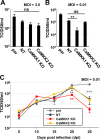
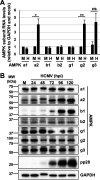
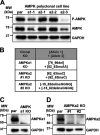
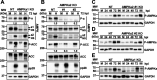
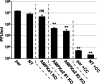
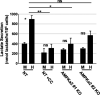
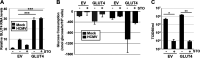
Human Cytomegalovirus (HCMV) infection modulates cellular metabolism to support viral replication. Calcium/calmodulin-dependent kinase kinase (CaMKK) and AMP-activated protein kinase (AMPK) regulate metabolic activation and have been found to be important for successful HCMV infection. Here, we explored the contributions that specific CaMKK isoforms and AMPK subunit isoforms make toward HCMV infection. Our results indicate that various CaMKK and AMPK isoforms contribute to infection in unique ways. For example, CaMKK1 is important for HCMV infection at a low multiplicity of infection, but is dispensable for AMPK activation at the earliest times of infection, which our data suggest is more reliant on CaMKK2. Our results also indicate that HCMV specifically induces the expression of the non-ubiquitous AMPKa2 catalytic subunit, found to be important for both HCMV-mediated glycolytic activation and high titer infection. Further, we find that AMPK-mediated glycolytic activation is important for infection, as overexpression of GLUT4, the high capacity glucose transporter, partially rescues viral replication in the face of AMPK inhibition. Collectively, our data indicate that HCMV infection selectively induces the expression of specific metabolic regulatory kinases, relying on their activity to support glycolytic activation and productive infection.IMPORTANCE Viruses are obligate parasites that depend on the host cell to provide the energy and molecular building blocks to mass produce infectious viral progeny. The processes that govern viral modulation of cellular resources have emerged as critical for successful infection. Here, we find that HCMV depends on two kinase isoforms to support infection, CaMKK1 and AMPKa2. We find that HCMV specifically induces expression of the AMPKa2 subunit to induce metabolic activation and drive robust viral replication. These results suggest that HCMV has evolved mechanisms to target specific metabolic regulatory kinase subunits to support productive infection, thereby providing insight into how HCMV hijacks cellular metabolism for its replication, and sheds light on potential viral therapeutic vulnerabilities.
Copyright © 2020 American Society for Microbiology.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

