This is a preprint.
Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020
- PMID: 33269368
- PMCID: PMC7709189
- DOI: 10.1101/2020.10.25.20219063
Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020
Update in
-
Spread of a SARS-CoV-2 variant through Europe in the summer of 2020.Nature. 2021 Jul;595(7869):707-712. doi: 10.1038/s41586-021-03677-y. Epub 2021 Jun 7. Nature. 2021. PMID: 34098568
Abstract
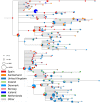
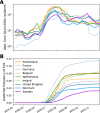
Following its emergence in late 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic resulting in unprecedented efforts to reduce transmission and develop therapies and vaccines (WHO Emergency Committee, 2020; Zhu et al., 2020). Rapidly generated viral genome sequences have allowed the spread of the virus to be tracked via phylogenetic analysis (Worobey et al., 2020; Hadfield et al., 2018; Pybus et al., 2020). While the virus spread globally in early 2020 before borders closed, intercontinental travel has since been greatly reduced, allowing continent-specific variants to emerge. However, within Europe travel resumed in the summer of 2020, and the impact of this travel on the epidemic is not well understood. Here we report on a novel SARS-CoV-2 variant, 20E (EU1), that emerged in Spain in early summer, and subsequently spread to multiple locations in Europe. We find no evidence of increased transmissibility of this variant, but instead demonstrate how rising incidence in Spain, resumption of travel across Europe, and lack of effective screening and containment may explain the variant's success. Despite travel restrictions and quarantine requirements, we estimate 20E (EU1) was introduced hundreds of times to countries across Europe by summertime travellers, likely undermining local efforts to keep SARS-CoV-2 cases low. Our results demonstrate how a variant can rapidly become dominant even in absence of a substantial transmission advantage in favorable epidemiological settings. Genomic surveillance is critical to understanding how travel can impact SARS-CoV-2 transmission, and thus for informing future containment strategies as travel resumes.
Conflict of interest statement
Transparency declaration DV is a consultant for Vir Biotechnology Inc. DC is an employee of Vir Biotechnology and may hold shares in Vir Biotechnology. The Veesler laboratory has received an unrelated sponsored research agreement from Vir Biotechnology Inc. AH is a co-founder of Kido Dynamics, DM is employed by Kido Dynamics. The other authors declare no competing interests.
Figures


References
-
- WHO Emergency Committee, Statement on the second meeting of the international health regulations (2005) emergency committee regarding the outbreak of novel coronavirus (2019-ncov), 2020.
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., Brief Report: A Novel Coronavirus from Patients with Pneumonia in China, 2019, The New England Journal of Medicine 382 (2020) 727. Publisher: NEJM Group. - PMC - PubMed
-
- Worobey M., Pekar J., Larsen B. B., Nelson M. I., Hill V., Joy J. B., Rambaut A., Suchard M. A., Wertheim J. O., Lemey P., The emergence of SARS-CoV-2 in Europe and North America, Science (2020). Publisher: American Association for the Advancement of Science Section: Research Article. - PMC - PubMed
-
- Pybus O., Rambaut A., et al. , Preliminary analysis of SARS-CoV-2 importation & establishment of UK transmission lineages, 2020.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
