Reconstitution of a mini-gene cluster combined with ribosome engineering led to effective enhancement of salinomycin production in Streptomyces albus
- PMID: 33270372
- PMCID: PMC8601195
- DOI: 10.1111/1751-7915.13686
Reconstitution of a mini-gene cluster combined with ribosome engineering led to effective enhancement of salinomycin production in Streptomyces albus
Abstract
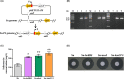
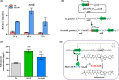
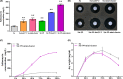
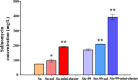
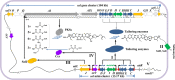
Salinomycin, an FDA-approved polyketide drug, was recently identified as a promising anti-tumour and anti-viral lead compound. It is produced by Streptomyces albus, and the biosynthetic gene cluster (sal) spans over 100 kb. The genetic manipulation of large polyketide gene clusters is challenging, and approaches delivering reliable efficiency and accuracy are desired. Herein, a delicate strategy to enhance salinomycin production was devised and evaluated. We reconstructed a minimized sal gene cluster (mini-cluster) on pSET152 including key genes responsible for tailoring modification, antibiotic resistance, positive regulation and precursor supply. These genes were overexpressed under the control of constitutive promoter PkasO* or Pneo . The pks operon was not included in the mini-cluster, but it was upregulated by SalJ activation. After the plasmid pSET152::mini-cluster was introduced into the wild-type strain and a chassis host strain obtained by ribosome engineering, salinomycin production was increased to 2.3-fold and 5.1-fold compared with that of the wild-type strain respectively. Intriguingly, mini-cluster introduction resulted in much higher production than overexpression of the whole sal gene cluster. The findings demonstrated that reconstitution of sal mini-cluster combined with ribosome engineering is an efficient novel approach and may be extended to other large polyketide biosynthesis.
© 2020 The Authors. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Conversion of the high-yield salinomycin producer Streptomyces albus BK3-25 into a surrogate host for polyketide production.Sci China Life Sci. 2017 Sep;60(9):1000-1009. doi: 10.1007/s11427-017-9122-8. Epub 2017 Aug 14. Sci China Life Sci. 2017. PMID: 28812299
-
Mechanism of salinomycin overproduction in Streptomyces albus as revealed by comparative functional genomics.Appl Microbiol Biotechnol. 2017 Jun;101(11):4635-4644. doi: 10.1007/s00253-017-8278-5. Epub 2017 Apr 11. Appl Microbiol Biotechnol. 2017. PMID: 28401259
-
Establishing an efficient salinomycin biosynthetic pathway in three heterologous Streptomyces hosts by constructing a 106-kb multioperon artificial gene cluster.Biotechnol Bioeng. 2021 Dec;118(12):4668-4677. doi: 10.1002/bit.27928. Epub 2021 Sep 1. Biotechnol Bioeng. 2021. PMID: 34436784
-
Biosynthesis, regulation, and engineering of a linear polyketide tautomycetin: a novel immunosuppressant in Streptomyces sp. CK4412.J Ind Microbiol Biotechnol. 2017 May;44(4-5):555-561. doi: 10.1007/s10295-016-1847-2. Epub 2016 Oct 12. J Ind Microbiol Biotechnol. 2017. PMID: 27734184 Review.
-
Herboxidiene biosynthesis, production, and structural modifications: prospect for hybrids with related polyketide.Appl Microbiol Biotechnol. 2015 Oct;99(20):8351-62. doi: 10.1007/s00253-015-6860-2. Epub 2015 Aug 19. Appl Microbiol Biotechnol. 2015. PMID: 26286508 Review.
Cited by
-
Transcriptome-guided identification of a four-component system, SbrH1-R, that modulates milbemycin biosynthesis by influencing gene cluster expression, precursor supply, and antibiotic efflux.Synth Syst Biotechnol. 2022 Feb 20;7(2):705-717. doi: 10.1016/j.synbio.2022.02.003. eCollection 2022 Jun. Synth Syst Biotechnol. 2022. PMID: 35261928 Free PMC article.
-
Synthetic Biology Tools for Engineering Microbial Cells to Fight Superbugs.Front Bioeng Biotechnol. 2022 May 4;10:869206. doi: 10.3389/fbioe.2022.869206. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35600895 Free PMC article. Review.
-
CRISPR/Cas9-Mediated Multi-Locus Promoter Engineering in ery Cluster to Improve Erythromycin Production in Saccharopolyspora erythraea.Microorganisms. 2023 Feb 28;11(3):623. doi: 10.3390/microorganisms11030623. Microorganisms. 2023. PMID: 36985197 Free PMC article.
-
Metabolic perturbation of Streptomyces albulus by introducing NADP-dependent glyceraldehyde 3-phosphate dehydrogenase.Front Microbiol. 2024 Jan 24;15:1328321. doi: 10.3389/fmicb.2024.1328321. eCollection 2024. Front Microbiol. 2024. PMID: 38328422 Free PMC article.
-
Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Deep-Sea Hydrothermal Vent-Derived Streptomyces sp. SCSIO ZS0520.Mar Drugs. 2022 Jun 14;20(6):393. doi: 10.3390/md20060393. Mar Drugs. 2022. PMID: 35736196 Free PMC article.
References
-
- Brautaset, T. , Sekurova, O.N. , Sletta, H. , Ellingsen, T.E. , StrŁm, A.R. , Valla, S. , and Zotchev, S.B. (2000) Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol 7: 395–403. - PubMed
-
- Caffrey, P. , Lynch, S. , Flood, E. , Finnan, S. , and Oliynyk, M. (2001) Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem Biol 8: 713–723. - PubMed
-
- Du, D. , Zhu, Y. , Wei, J. , Tian, Y. , Niu, G. , and Tan, H. (2013) Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl Microbiol Biotechnol 97: 6383–6396. - PubMed
-
- Gibson, D.G. , Glass, J.I. , Lartigue, C. , Noskov, V.N. , Chuang, R.Y. , Algire, M.A. , et al. (2010) Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329: 52–56. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

