Complex genetic dependencies among growth and neurological phenotypes in healthy children: Towards deciphering developmental mechanisms
- PMID: 33270668
- PMCID: PMC7714163
- DOI: 10.1371/journal.pone.0242684
Complex genetic dependencies among growth and neurological phenotypes in healthy children: Towards deciphering developmental mechanisms
Abstract
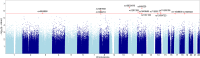
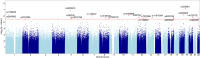
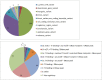
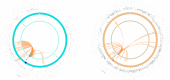
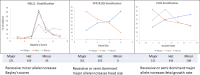
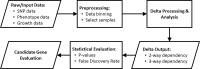
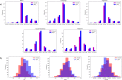
The genetic mechanisms of childhood development in its many facets remain largely undeciphered. In the population of healthy infants studied in the Growing Up in Singapore Towards Healthy Outcomes (GUSTO) program, we have identified a range of dependencies among the observed phenotypes of fetal and early childhood growth, neurological development, and a number of genetic variants. We have quantified these dependencies using our information theory-based methods. The genetic variants show dependencies with single phenotypes as well as pleiotropic effects on more than one phenotype and thereby point to a large number of brain-specific and brain-expressed gene candidates. These dependencies provide a basis for connecting a range of variants with a spectrum of phenotypes (pleiotropy) as well as with each other. A broad survey of known regulatory expression characteristics, and other function-related information from the literature for these sets of candidate genes allowed us to assemble an integrated body of evidence, including a partial regulatory network, that points towards the biological basis of these general dependencies. Notable among the implicated loci are RAB11FIP4 (next to NF1), MTMR7 and PLD5, all highly expressed in the brain; DNMT1 (DNA methyl transferase), highly expressed in the placenta; and PPP1R12B and DMD (dystrophin), known to be important growth and development genes. While we cannot specify and decipher the mechanisms responsible for the phenotypes in this study, a number of connections for further investigation of fetal and early childhood growth and neurological development are indicated. These results and this approach open the door to new explorations of early human development.
Conflict of interest statement
The authors have read the journal's policy and have the following competing interests: JDW and JLF are paid employees of Metrum Research Group. NLJ is a paid employee of Pharmactuarials LLC. There are no patents, products in development or marketed products to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

