Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression
- PMID: 33270893
- PMCID: PMC7736780
- DOI: 10.1093/nar/gkaa1151
Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression
Abstract
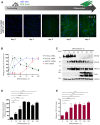
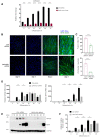
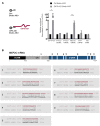
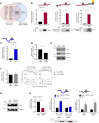
Long noncoding (lnc)RNAs potently regulate gene expression programs in physiology and disease. Here, we describe a key function for lncRNA OIP5-AS1 in myogenesis, the process whereby myoblasts differentiate into myotubes during muscle development and muscle regeneration after injury. In human myoblasts, OIP5-AS1 levels increased robustly early in myogenesis, and its loss attenuated myogenic differentiation and potently reduced the levels of the myogenic transcription factor MEF2C. This effect relied upon the partial complementarity of OIP5-AS1 with MEF2C mRNA and the presence of HuR, an RNA-binding protein (RBP) with affinity for both transcripts. Remarkably, HuR binding to MEF2C mRNA, which stabilized MEF2C mRNA and increased MEF2C abundance, was lost after OIP5-AS1 silencing, suggesting that OIP5-AS1 might serve as a scaffold to enhance HuR binding to MEF2C mRNA, in turn increasing MEF2C production. These results highlight a mechanism whereby a lncRNA promotes myogenesis by enhancing the interaction of an RBP and a myogenic mRNA.
Published by Oxford University Press on behalf of Nucleic Acids Research 2020.
Figures

References
-
- Baracos V.E., Martin L., Korc M., Guttridge D.C., Fearon K.C.H.. Cancer-associated cachexia. Nature Rev. 2018; 4:17105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

