Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency
- PMID: 33271069
- PMCID: PMC7939546
- DOI: 10.1016/j.stem.2020.11.005
Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency
Erratum in
-
Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency.Cell Stem Cell. 2021 Dec 2;28(12):2180. doi: 10.1016/j.stem.2021.11.002. Cell Stem Cell. 2021. PMID: 34861148 Free PMC article. No abstract available.
Abstract
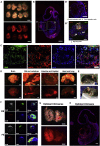
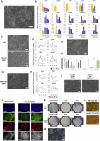
Pluripotent cells emerge as a naive founder population in the blastocyst, acquire capacity for germline and soma formation, and then undergo lineage priming. Mouse embryonic stem cells (ESCs) and epiblast-derived stem cells (EpiSCs) represent the initial naive and final primed phases of pluripotency, respectively. Here, we investigate the intermediate formative stage. Using minimal exposure to specification cues, we derive stem cells from formative mouse epiblast. Unlike ESCs or EpiSCs, formative stem (FS) cells respond directly to germ cell induction. They colonize somatic tissues and germline in chimeras. Whole-transcriptome analyses show similarity to pre-gastrulation formative epiblast. Signal responsiveness and chromatin accessibility features reflect lineage capacitation. Furthermore, FS cells show distinct transcription factor dependencies, relying critically on Otx2. Finally, FS cell culture conditions applied to human naive cells or embryos support expansion of similar stem cells, consistent with a conserved staging post on the trajectory of mammalian pluripotency.
Keywords: chimaera; epiblast; formative pluripotency; lineage induction; pluripotent stem cell; primordial germ cell; self-renewal.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests
Figures






Comment in
-
Stabilizing Formative Pluripotent States with Germ Cell Competency.Cell Stem Cell. 2021 Mar 4;28(3):361-363. doi: 10.1016/j.stem.2021.02.021. Cell Stem Cell. 2021. PMID: 33667353
References
-
- Acampora D., Di Giovannantonio L.G., Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140:43–55. - PubMed
-
- Acampora D., Omodei D., Petrosino G., Garofalo A., Savarese M., Nigro V., Di Giovannantonio L.G., Mercadante V., Simeone A. Loss of the Otx2-Binding Site in the Nanog Promoter Affects the Integrity of Embryonic Stem Cell Subtypes and Specification of Inner Cell Mass-Derived Epiblast. Cell Rep. 2016;15:2651–2664. - PubMed
-
- Allison T.F., Smith A.J.H., Anastassiadis K., Sloane-Stanley J., Biga V., Stavish D., Hackland J., Sabri S., Langerman J., Jones M. Identification and Single-Cell Functional Characterization of an Endodermally Biased Pluripotent Substate in Human Embryonic Stem Cells. Stem Cell Reports. 2018;10:1895–1907. - PMC - PubMed
-
- Ang S.L., Jin O., Rhinn M., Daigle N., Stevenson L., Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

