Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses
- PMID: 33271916
- PMCID: PMC7761488
- DOI: 10.3390/v12121376
Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses
Abstract
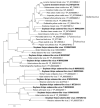
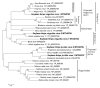
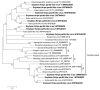
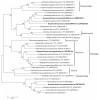
Soybean thrips (Neohydatothrips variabilis) are one of the most efficient vectors of soybean vein necrosis virus, which can cause severe necrotic symptoms in sensitive soybean plants. To determine which other viruses are associated with soybean thrips, the metatranscriptome of soybean thrips, collected by the Midwest Suction Trap Network during 2018, was analyzed. Contigs assembled from the data revealed a remarkable diversity of virus-like sequences. Of the 181 virus-like sequences identified, 155 were novel and associated primarily with taxa of arthropod-infecting viruses, but sequences similar to plant and fungus-infecting viruses were also identified. The novel viruses were predicted to have positive-sense RNA, negative-stranded RNA, double-stranded RNA, and single-stranded DNA genomes. The assembled sequences included 100 contigs that represented at least 95% coverage of a virus genome or genome segment. Sequences represented 12 previously described arthropod viruses including eight viruses reported from Hubei Province in China, and 12 plant virus sequences of which six have been previously described. The presence of diverse populations of plant viruses within soybean thrips suggests they feed on and acquire viruses from multiple host plant species that could be transmitted to soybean. Assessment of the virome of soybean thrips provides, for the first time, information on the diversity of viruses present in thrips.
Keywords: Neohydatothrips variabilis; soybean thrips; vector-enabled transcriptomics; viral metatranscriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures












References
-
- Jones D.R. Plant viruses transmitted by thrips. Eur. J. Plant Pathol. 2005;113:119–157. doi: 10.1007/s10658-005-2334-1. - DOI
-
- Irizarry M.D., Elmore M.G., Batzer J.C., Whitham S.A., Mueller D.S. Alternative hosts for soybean vein necrosis virus and feeding preferences of its vector soybean thrips. Plant Health Prog. 2018;19:176–181. doi: 10.1094/PHP-11-17-0071-RS. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

