RB Regulates DNA Double Strand Break Repair Pathway Choice by Mediating CtIP Dependent End Resection
- PMID: 33271982
- PMCID: PMC7730402
- DOI: 10.3390/ijms21239176
RB Regulates DNA Double Strand Break Repair Pathway Choice by Mediating CtIP Dependent End Resection
Abstract
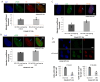
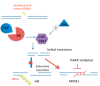
Inactivation of the retinoblastoma tumor suppressor gene (RB1) leads to genome instability, and can be detected in retinoblastoma and other cancers. One damaging effect is causing DNA double strand breaks (DSB), which, however, can be repaired by homologous recombination (HR), classical non-homologous end joining (C-NHEJ), and micro-homology mediated end joining (MMEJ). We aimed to study the mechanistic roles of RB in regulating multiple DSB repair pathways. Here we show that HR and C-NHEJ are decreased, but MMEJ is elevated in RB-depleted cells. After inducing DSB by camptothecin, RB co-localizes with CtIP, which regulates DSB end resection. RB depletion leads to less RPA and native BrdU foci, which implies less end resection. In RB-depleted cells, less CtIP foci, and a lack of phosphorylation on CtIP Thr847, are observed. According to the synthetic lethality principle, based on the altered DSB repair pathway choice, after inducing DSBs by camptothecin, RB depleted cells are more sensitive to co-treatment with camptothecin and MMEJ blocker poly-ADP ribose polymerase 1 (PARP1) inhibitor. We propose a model whereby RB can regulate DSB repair pathway choice by mediating the CtIP dependent DNA end resection. The use of PARP1 inhibitor could potentially improve treatment outcomes for RB-deficient cancers.
Keywords: CtIP; RB; classical non-homologous end joining; homologous recombination; micro-homology mediated end joining; resection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Nicolay B.N., Danielian P.S., Kottakis F., Lapek J.D., Jr., Sanidas I., Miles W.O., Dehnad M., Tschöp K., Gierut J.J., Manning A.L., et al. Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev. 2015;29:1875–1889. doi: 10.1101/gad.264127.115. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

