A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings
- PMID: 33271988
- PMCID: PMC7731377
- DOI: 10.3390/ijms21239177
A Long-Read Sequencing Approach for Direct Haplotype Phasing in Clinical Settings
Abstract
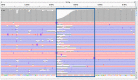
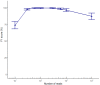
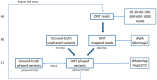
The reconstruction of individual haplotypes can facilitate the interpretation of disease risks; however, high costs and technical challenges still hinder their assessment in clinical settings. Second-generation sequencing is the gold standard for variant discovery but, due to the production of short reads covering small genomic regions, allows only indirect haplotyping based on statistical methods. In contrast, third-generation methods such as the nanopore sequencing platform developed by Oxford Nanopore Technologies (ONT) generate long reads that can be used for direct haplotyping, with fewer drawbacks. However, robust standards for variant phasing in ONT-based target resequencing efforts are not yet available. In this study, we presented a streamlined proof-of-concept workflow for variant calling and phasing based on ONT data in a clinically relevant 12-kb region of the APOE locus, a hotspot for variants and haplotypes associated with aging-related diseases and longevity. Starting with sequencing data from simple amplicons of the target locus, we demonstrated that ONT data allow for reliable single-nucleotide variant (SNV) calling and phasing from as little as 60 reads, although the recognition of indels is less efficient. Even so, we identified the best combination of ONT read sets (600) and software (BWA/Minimap2 and HapCUT2) that enables full haplotype reconstruction when both SNVs and indels have been identified previously using a highly-accurate sequencing platform. In conclusion, we established a rapid and inexpensive workflow for variant phasing based on ONT long reads. This allowed for the analysis of multiple samples in parallel and can easily be implemented in routine clinical practice, including diagnostic testing.
Keywords: diagnostic testing; haplotype phasing; nanopore sequencing; variant calling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Scitable by Nature Education. [(accessed on 30 November 2020)]; Available online: https://www.nature.com/scitable/definition/haplotype-haplotypes-142/
-
- Allen M., Kachadoorian M., Quicksall Z., Zou F., Chai H.S., Younkin C., E Crook J., Pankratz V.S., Carrasquillo M.M., Krishnan S., et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimer’s Res. Ther. 2014;6:39. doi: 10.1186/alzrt268. - DOI - PMC - PubMed
-
- Lescai F., Chiamenti A.M., Codemo A., Pirazzini C., D’Agostino G., Ruaro C., Ghidoni R., Benussi L., Galimberti D., Esposito F., et al. An APOE Haplotype Associated with Decreased epsilon4 Expression Increases the Risk of Late Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2011;24:235–245. doi: 10.3233/JAD-2011-101764. - DOI - PubMed
-
- Navarro S., Medina P., Mira Y., Estelles A., Villa P., Ferrando F., Vaya A., Bertina R.M., España F. Haplotypes of the EPCR gene, prothrombin levels, and the risk of venous thrombosis in carriers of the prothrombin G20210A mutation. Haematol. 2008;93:885–891. doi: 10.3324/haematol.12448. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

