Local environment effects on charged mutations for developing aggregation-resistant monoclonal antibodies
- PMID: 33273506
- PMCID: PMC7713239
- DOI: 10.1038/s41598-020-78136-1
Local environment effects on charged mutations for developing aggregation-resistant monoclonal antibodies
Abstract
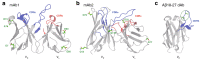
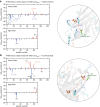
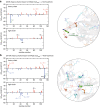
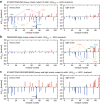
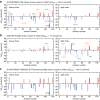
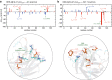
Protein aggregation is a major concern in biotherapeutic applications of monoclonal antibodies. Introducing charged mutations is among the promising strategies to improve aggregation resistance. However, the impact of such mutations on solubilizing activity depends largely on the inserting location, whose mechanism is still not well understood. Here, we address this issue from a solvation viewpoint, and this is done by analyzing how the change in solvation free energy upon charged mutation is composed of individual contributions from constituent residues. To this end, we perform molecular dynamics simulations for a number of antibody mutants and carry out the residue-wise decomposition of the solvation free energy. We find that, in addition to the previously identified "global" principle emphasizing the key role played by the protein total net charge, a local net charge within [Formula: see text]15 Å from the mutation site exerts significant effects. For example, when the net charge of an antibody is positive, the global principle states that introducing a positively charged mutation will lead to more favorable solvation. Our finding further adds that an even more optimal mutation can be done at the site around which more positively charged residues and fewer negatively charged residues are present. Such a "local" design principle accounts for the location dependence of charged mutations, and will be useful in producing aggregation-resistant antibodies.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Optimal charged mutations in the complementarity-determining regions that prevent domain antibody aggregation are dependent on the antibody scaffold.Protein Eng Des Sel. 2014 Feb;27(2):29-39. doi: 10.1093/protein/gzt058. Epub 2014 Jan 6. Protein Eng Des Sel. 2014. PMID: 24398633
-
Rational design of viscosity reducing mutants of a monoclonal antibody: hydrophobic versus electrostatic inter-molecular interactions.MAbs. 2015;7(1):212-30. doi: 10.4161/19420862.2014.985504. MAbs. 2015. PMID: 25559441 Free PMC article.
-
Distinct role of hydration water in protein misfolding and aggregation revealed by fluctuating thermodynamics analysis.Acc Chem Res. 2015 Apr 21;48(4):956-65. doi: 10.1021/acs.accounts.5b00032. Epub 2015 Apr 6. Acc Chem Res. 2015. PMID: 25844814 Review.
-
Aggregation-resistant domain antibodies engineered with charged mutations near the edges of the complementarity-determining regions.Protein Eng Des Sel. 2012 Oct;25(10):591-601. doi: 10.1093/protein/gzs042. Epub 2012 Jul 27. Protein Eng Des Sel. 2012. PMID: 22843678
-
Alteration of Physicochemical Properties for Antibody-Drug Conjugates and Their Impact on Stability.J Pharm Sci. 2020 Jan;109(1):161-168. doi: 10.1016/j.xphs.2019.08.006. Epub 2019 Aug 10. J Pharm Sci. 2020. PMID: 31408634 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

