SIRT3, a metabolic target linked to ataxia-telangiectasia mutated (ATM) gene deficiency in diffuse large B-cell lymphoma
- PMID: 33273545
- PMCID: PMC7712916
- DOI: 10.1038/s41598-020-78193-6
SIRT3, a metabolic target linked to ataxia-telangiectasia mutated (ATM) gene deficiency in diffuse large B-cell lymphoma
Abstract
Inactivation of Ataxia-telangiectasia mutated (ATM) gene results in an increased risk to develop cancer. We show that ATM deficiency in diffuse large B-cell lymphoma (DLBCL) significantly induce mitochondrial deacetylase sirtuin-3 (SIRT3) activity, disrupted mitochondrial structure, decreased mitochondrial respiration, and compromised TCA flux compared with DLBCL cells expressing wild type (WT)-ATM. This corresponded to enrichment of glutamate receptor and glutamine pathways in ATM deficient background compared to WT-ATM DLBCL cells. ATM-/- DLBCL cells have decreased apoptosis in contrast to radiosensitive non-cancerous A-T cells. In vivo studies using gain and loss of SIRT3 expression showed that SIRT3 promotes growth of ATM CRISPR knockout DLBCL xenografts compared to wild-type ATM control xenografts. Importantly, screening of DLBCL patient samples identified SIRT3 as a putative therapeutic target, and validated an inverse relationship between ATM and SIRT3 expression. Our data predicts SIRT3 as an important therapeutic target for DLBCL patients with ATM null phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures
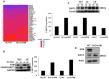


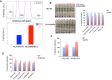
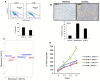

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

