Methods for the discovery of small molecules to monitor and perturb the activity of the human proteasome
- PMID: 33275045
- PMCID: PMC7857359
- DOI: 10.4155/fmc-2020-0288
Methods for the discovery of small molecules to monitor and perturb the activity of the human proteasome
Abstract
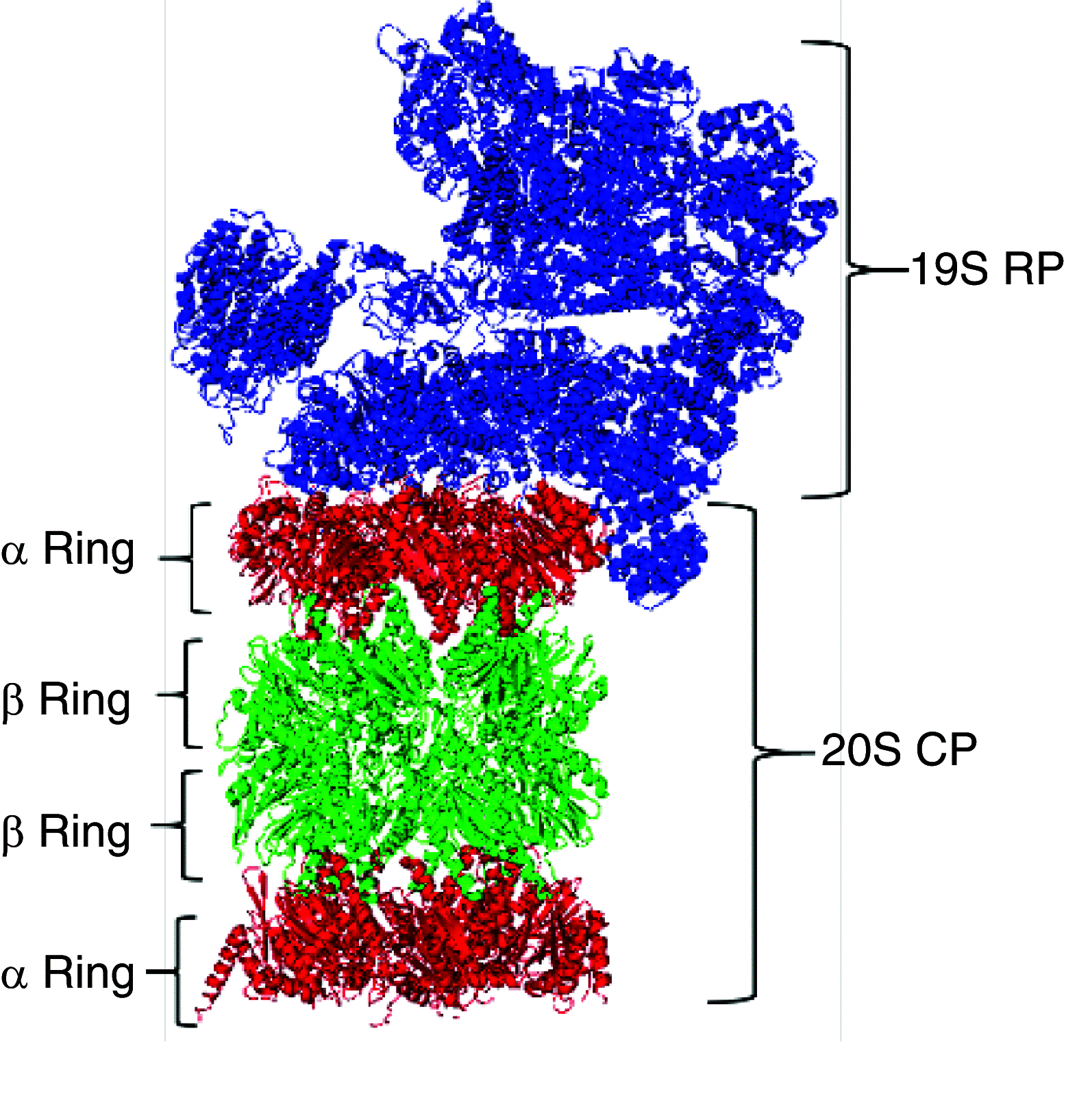
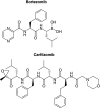
Regulating protein production and degradation is critical to maintaining cellular homeostasis. The proteasome is a key player in keeping proteins at the proper levels. However, proteasome activity can be altered in certain disease states, such as blood cancers and neurodegenerative diseases. Cancers often exhibit enhanced proteasomal activity, as protein synthesis is increased in these cells compared with normal cells. Conversely, neurodegenerative diseases are characterized by protein accumulation, leading to reduced proteasome activity. As a result, the proteasome has emerged as a target for therapeutic intervention. The potential of the proteasome as a therapeutic target has come from studies involving chemical stimulators and inhibitors, and the development of a suite of assays and probes that can be used to monitor proteasome activity with purified enzyme and in live cells.
Keywords: degradation; proteasome; screening.
Conflict of interest statement
This work was supported through the Purdue University Center for Cancer Research NIH grant (P30 CA023168), the American Cancer Society Institutional Research Grant (IRG-14-190-56) to the Purdue University Center for Cancer Research, and a grant from the NIH-NIGMS (R21GM131206). Prof. Trader is a scientific co-founder and stock holder of Booster Therapeutics GmbH. The other authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures

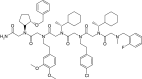

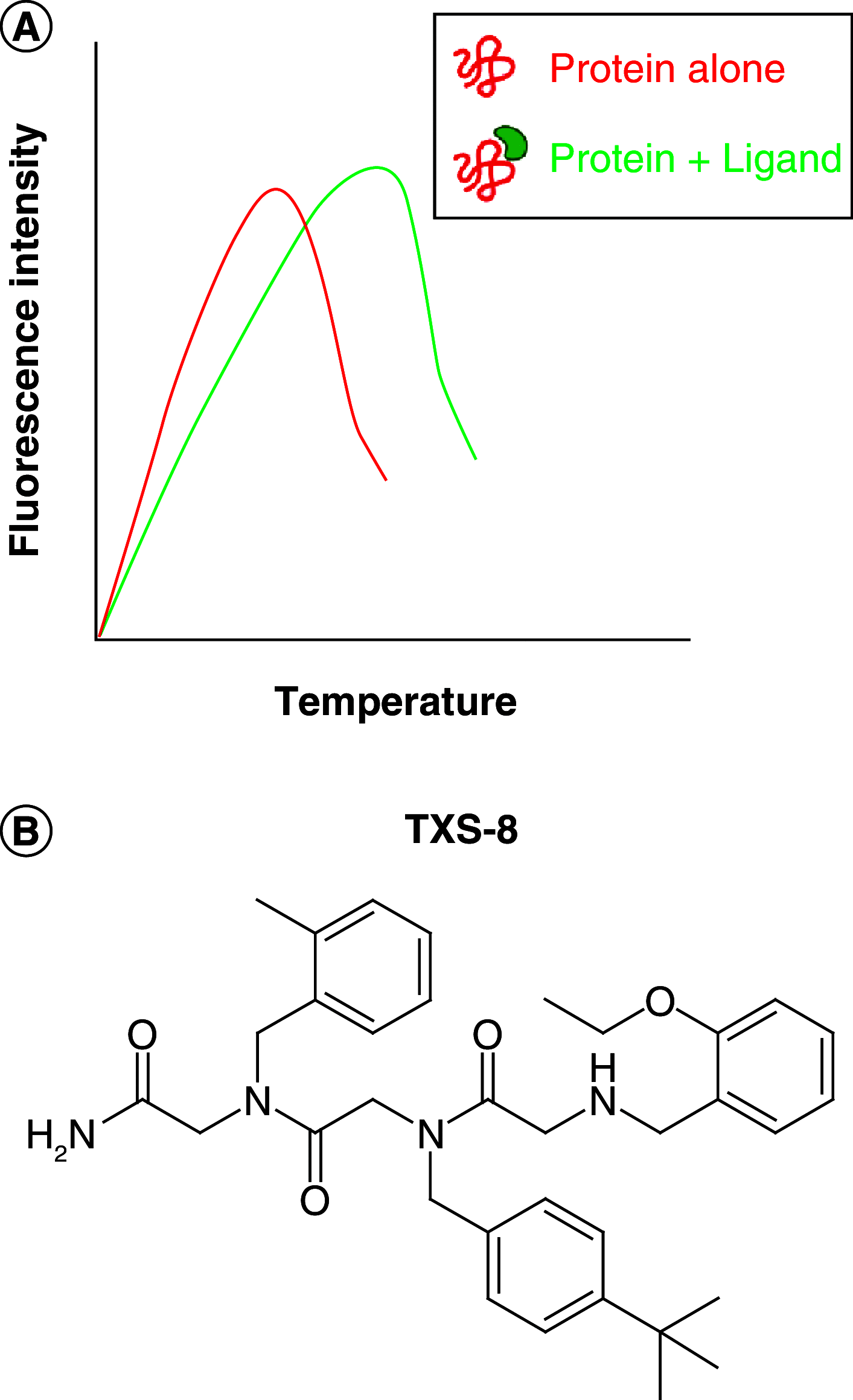
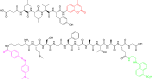

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
