A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody
- PMID: 33275640
- PMCID: PMC7744049
- DOI: 10.1371/journal.ppat.1009089
A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody
Abstract
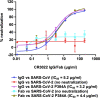
Epitopes that are conserved among SARS-like coronaviruses are attractive targets for design of cross-reactive vaccines and therapeutics. CR3022 is a SARS-CoV neutralizing antibody to a highly conserved epitope on the receptor binding domain (RBD) on the spike protein that is able to cross-react with SARS-CoV-2, but with lower affinity. Using x-ray crystallography, mutagenesis, and binding experiments, we illustrate that of four amino acid differences in the CR3022 epitope between SARS-CoV-2 and SARS-CoV, a single mutation P384A fully determines the affinity difference. CR3022 does not neutralize SARS-CoV-2, but the increased affinity to SARS-CoV-2 P384A mutant now enables neutralization with a similar potency to SARS-CoV. We further investigated CR3022 interaction with the SARS-CoV spike protein by negative-stain EM and cryo-EM. Three CR3022 Fabs bind per trimer with the RBD observed in different up-conformations due to considerable flexibility of the RBD. In one of these conformations, quaternary interactions are made by CR3022 to the N-terminal domain (NTD) of an adjacent subunit. Overall, this study provides insights into antigenic variation and potential cross-neutralizing epitopes on SARS-like viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Update of
-
A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody.bioRxiv [Preprint]. 2020 Sep 21:2020.09.21.305441. doi: 10.1101/2020.09.21.305441. bioRxiv. 2020. Update in: PLoS Pathog. 2020 Dec 4;16(12):e1009089. doi: 10.1371/journal.ppat.1009089. PMID: 32995788 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

