The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis
- PMID: 33276435
- PMCID: PMC7760870
- DOI: 10.3390/plants9121692
The Complete Chloroplast Genome of the Vulnerable Oreocharis esquirolii (Gesneriaceae): Structural Features, Comparative and Phylogenetic Analysis
Abstract
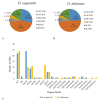
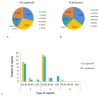
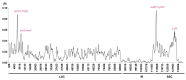
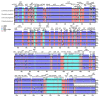
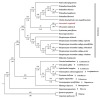
Oreocharis esquirolii, a member of Gesneriaceae, is known as Thamnocharis esquirolii, which has been regarded a synonym of the former. The species is endemic to Guizhou, southwestern China, and is evaluated as vulnerable (VU) under the International Union for Conservation of Nature (IUCN) criteria. Until now, the sequence and genome information of O. esquirolii remains unknown. In this study, we assembled and characterized the complete chloroplast (cp) genome of O. esquirolii using Illumina sequencing data for the first time. The total length of the cp genome was 154,069 bp with a typical quadripartite structure consisting of a pair of inverted repeats (IRs) of 25,392 bp separated by a large single copy region (LSC) of 85,156 bp and a small single copy region (SSC) of18,129 bp. The genome comprised 114 unique genes with 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Thirty-one repeat sequences and 74 simple sequence repeats (SSRs) were identified. Genome alignment across five plastid genomes of Gesneriaceae indicated a high sequence similarity. Four highly variable sites (rps16-trnQ, trnS-trnG, ndhF-rpl32, and ycf 1) were identified. Phylogenetic analysis indicated that O. esquirolii grouped together with O. mileensis, supporting resurrection of the name Oreocharis esquirolii from Thamnocharisesquirolii. The complete cp genome sequence will contribute to further studies in molecular identification, genetic diversity, and phylogeny.
Keywords: Gesneriaceae; Oreocharis; Thamnocharis; complete chloroplast genome; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bentham G. Gesneriaceae. In: Bentham G., Hooker J.D., editors. Genera Plantarum. Lovell Reeve & Co.; London, UK: 1876. pp. 990–1025.
-
- Wang W.T., Pan K.Y., Li Z.Y., Weitzman A.L., Skog L.E. Gesneriaceae. In: Wu C.Y., Raven P.H., Hong D.Y., editors. Flora of China. Volume 18. Science Press; Beijing, China: Missouri Botanical Garden Press; St. Louis, MO, USA: 1998. pp. 244–401.
-
- Wang Y.Z., Liang R.H., Wang B.H., Li J.M., Qiu Z.J., Li Z.Y., Weber A. Origin and phylogenetic relationships of the Old World Gesneriaceae with actinomorphic flowers inferred from ITS and trnL-trnF sequences. Taxon. 2010;59:1044–1052. doi: 10.1002/tax.594005. - DOI
-
- Möller M., Middleton D., Nishii K., Wei Y.G., Sontag S., Weber A. A new delineation for Oreocharis incorporating an additional ten genera of Chinese Gesneriaceae. Phytotaxa. 2011;23:1–36. doi: 10.11646/phytotaxa.23.1.1. - DOI
-
- Möller M., Forrest A., Wei Y.G., Weber A. A molecular phylogenetic assessment of the advanced Asiatic and Malesian didymocarpoid Gesneriaceae with focus on non-monophyletic and monotypic genera. Plant Syst. Evol. 2011;292:223–248. doi: 10.1007/s00606-010-0413-z. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

