Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus
- PMID: 33276730
- PMCID: PMC7716463
- DOI: 10.1186/s12870-020-02752-2
Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus
Abstract
Background: Transcription factors GATAs are involved in plant developmental processes and respond to environmental stresses through binding DNA regulatory regions to regulate their downstream genes. However, little information on the GATA genes in Brassica napus is available. The release of the reference genome of B. napus provides a good opportunity to perform a genome-wide characterization of GATA family genes in rapeseed.
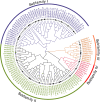
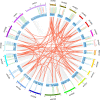
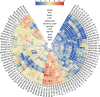
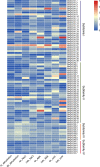
Results: In this study, 96 GATA genes randomly distributing on 19 chromosomes were identified in B. napus, which were classified into four subfamilies based on phylogenetic analysis and their domain structures. The amino acids of BnGATAs were obvious divergence among four subfamilies in terms of their GATA domains, structures and motif compositions. Gene duplication and synteny between the genomes of B. napus and A. thaliana were also analyzed to provide insights into evolutionary characteristics. Moreover, BnGATAs showed different expression patterns in various tissues and under diverse abiotic stresses. Single nucleotide polymorphisms (SNPs) distributions of BnGATAs in a core collection germplasm are probably associated with functional disparity under environmental stress condition in different genotypes of B. napus.
Conclusion: The present study was investigated genomic structures, evolution features, expression patterns and SNP distributions of 96 BnGATAs. The results enrich our understanding of the GATA genes in rapeseed.
Keywords: Brassica napus; Expression patterns; GATA; Genome-wide; SNP distribution.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Yuan Q, Zhang C, Zhao T, Yao M, Xu X. A genome-wide analysis of GATA transcription factor family in tomato and analysis of expression patterns. Int J Agric Biol. 2018;20(6):1274–1282.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

