RNA sequencing: new technologies and applications in cancer research
- PMID: 33276803
- PMCID: PMC7716291
- DOI: 10.1186/s13045-020-01005-x
RNA sequencing: new technologies and applications in cancer research
Abstract
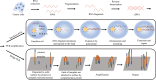
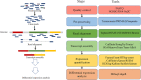
Over the past few decades, RNA sequencing has significantly progressed, becoming a paramount approach for transcriptome profiling. The revolution from bulk RNA sequencing to single-molecular, single-cell and spatial transcriptome approaches has enabled increasingly accurate, individual cell resolution incorporated with spatial information. Cancer, a major malignant and heterogeneous lethal disease, remains an enormous challenge in medical research and clinical treatment. As a vital tool, RNA sequencing has been utilized in many aspects of cancer research and therapy, including biomarker discovery and characterization of cancer heterogeneity and evolution, drug resistance, cancer immune microenvironment and immunotherapy, cancer neoantigens and so on. In this review, the latest studies on RNA sequencing technology and their applications in cancer are summarized, and future challenges and opportunities for RNA sequencing technology in cancer applications are discussed.
Keywords: Application; Cancer; RNA sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

