ATG5-Dependent Autophagy Uncouples T-cell Proliferative and Effector Functions and Separates Graft-versus-Host Disease from Graft-versus-Leukemia
- PMID: 33277367
- PMCID: PMC9400550
- DOI: 10.1158/0008-5472.CAN-20-1346
ATG5-Dependent Autophagy Uncouples T-cell Proliferative and Effector Functions and Separates Graft-versus-Host Disease from Graft-versus-Leukemia
Abstract
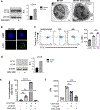
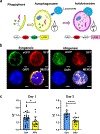
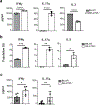
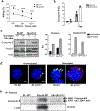
Autophagy is a vital cellular process whose role in T immune cells is poorly understood, specifically, in its regulation of allo-immunity. Stimulation of wild-type T cells in vitro and in vivo with allo-antigens enhances autophagy. To assess the relevance of autophagy to T-cell allo-immunity, we generated T-cell-specific Atg5 knock-out mice. Deficiency of ATG5-dependent autophagy reduced T-cell proliferation and increased apoptosis following in vitro and in vivo allo-stimulation. The absence of ATG5 in allo-stimulated T cells enhanced their ability to release effector cytokines and cytotoxic functions, uncoupling their proliferation and effector functions. Absence of autophagy reduced intracellular degradation of cytotoxic enzymes such as granzyme B, thus enhancing the cytotoxicity of T cells. In several in vivo models of allo-HSCT, ATG5-dependent dissociation of T-cell functions contributed to significant reduction in graft-versus-host disease (GVHD) but retained sufficient graft versus tumor (GVT) response. Our findings demonstrate that ATG5-dependent autophagy uncouples T-cell proliferation from its effector functions and offers a potential new strategy to enhance outcomes after allo-HSCT. SIGNIFICANCE: These findings demonstrate that induction of autophagy in donor T-cell promotes GVHD, while inhibition of T-cell autophagy mitigates GVHD without substantial loss of GVL responses.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

