Host genetics and infectious disease: new tools, insights and translational opportunities
- PMID: 33277640
- PMCID: PMC7716795
- DOI: 10.1038/s41576-020-00297-6
Host genetics and infectious disease: new tools, insights and translational opportunities
Abstract
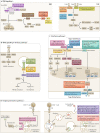
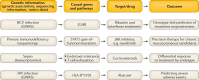
Understanding how human genetics influence infectious disease susceptibility offers the opportunity for new insights into pathogenesis, potential drug targets, risk stratification, response to therapy and vaccination. As new infectious diseases continue to emerge, together with growing levels of antimicrobial resistance and an increasing awareness of substantial differences between populations in genetic associations, the need for such work is expanding. In this Review, we illustrate how our understanding of the host-pathogen relationship is advancing through holistic approaches, describing current strategies to investigate the role of host genetic variation in established and emerging infections, including COVID-19, the need for wider application to diverse global populations mirroring the burden of disease, the impact of pathogen and vector genetic diversity and a broad array of immune and inflammation phenotypes that can be mapped as traits in health and disease. Insights from study of inborn errors of immunity and multi-omics profiling together with developments in analytical methods are further advancing our knowledge of this important area.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Stanaway JD, et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1923–1994. doi: 10.1016/S0140-6736(18)32225-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

