2-Pyridone natural products as inhibitors of SARS-CoV-2 main protease
- PMID: 33278462
- PMCID: PMC7710351
- DOI: 10.1016/j.cbi.2020.109348
2-Pyridone natural products as inhibitors of SARS-CoV-2 main protease
Abstract
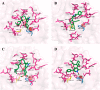
The disease, COVID-19, is caused by the severe acute respiratory coronavirus 2 (SARS-CoV-2) for which there is currently no treatment. The SARS-CoV-2 main protease (Mpro) is an important enzyme for viral replication. Small molecules that inhibit this protease could lead to an effective COVID-19 treatment. The 2-pyridone scaffold was previously identified as a possible key pharmacophore to inhibit SARS-CoV-2 Mpro. A search for natural, antimicrobial products with the 2-pyridone moiety was undertaken herein, and their calculated potency as inhibitors of SARS-CoV-2 Mpro was investigated. Thirty-three natural products containing the 2-pyridone scaffold were identified from the literature. An in silico methodology using AutoDock was employed to predict the binding energies and inhibition constants (Ki values) for each 2-pyridone-containing compound with SARS-CoV-2 Mpro. This consisted of molecular optimization of the 2-pyridone compound, docking of the compound with a crystal structure of SARS-CoV-2 Mpro, and evaluation of the predicted interactions and ligand-enzyme conformations. All compounds investigated bound to the active site of SARS-CoV-2 Mpro, close to the catalytic dyad (His-41 and Cys-145). Thirteen molecules had predicted Ki values <1 μM. Glu-166 formed a key hydrogen bond in the majority of the predicted complexes, while Met-165 had some involvement in the complex binding as a close contact to the ligand. Prominent 2-pyridone compounds were further evaluated for their ADMET properties. This work has identified 2-pyridone natural products with calculated potent inhibitory activity against SARS-CoV-2 Mpro and with desirable drug-like properties, which may lead to the rapid discovery of a treatment for COVID-19.
Keywords: 2-Pyridone; AutoDock; COVID-19; In silico molecular modelling; Main protease (M(pro)); Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors.Bioorg Med Chem. 2021 Jan 1;29:115860. doi: 10.1016/j.bmc.2020.115860. Epub 2020 Nov 6. Bioorg Med Chem. 2021. PMID: 33191083 Free PMC article. Review.
-
Establishing an Analogue Based In Silico Pipeline in the Pursuit of Novel Inhibitory Scaffolds against the SARS Coronavirus 2 Papain-Like Protease.Molecules. 2021 Feb 20;26(4):1134. doi: 10.3390/molecules26041134. Molecules. 2021. PMID: 33672721 Free PMC article.
-
In silico analysis of selected alkaloids against main protease (Mpro) of SARS-CoV-2.Chem Biol Interact. 2020 Dec 1;332:109309. doi: 10.1016/j.cbi.2020.109309. Epub 2020 Nov 9. Chem Biol Interact. 2020. PMID: 33181114 Free PMC article.
-
Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.Biochemistry. 2021 Oct 5;60(39):2925-2931. doi: 10.1021/acs.biochem.1c00414. Epub 2021 Sep 10. Biochemistry. 2021. PMID: 34506130 Free PMC article.
-
An Updated Review on SARS-CoV-2 Main Proteinase (MPro): Protein Structure and Small-Molecule Inhibitors.Curr Top Med Chem. 2021;21(6):442-460. doi: 10.2174/1568026620666201207095117. Curr Top Med Chem. 2021. PMID: 33292134 Review.
Cited by
-
Synthesis of a Pyridone-Based Phthalimide Fleximer and Its Characterization and Supramolecular Property Evaluation.ACS Omega. 2022 Jul 7;7(28):24485-24497. doi: 10.1021/acsomega.2c02095. eCollection 2022 Jul 19. ACS Omega. 2022. PMID: 35874266 Free PMC article.
-
Synthesis and Biological Evaluation of Benzothiazolyl-pyridine Hybrids as New Antiviral Agents against H5N1 Bird Flu and SARS-COV-2 Viruses.ACS Omega. 2023 Sep 25;8(40):36636-36654. doi: 10.1021/acsomega.3c01987. eCollection 2023 Oct 10. ACS Omega. 2023. PMID: 37841136 Free PMC article.
-
Phenothiazine-linked glutamic acid dendrons: an easy access and a new class of SARS-CoV-2 main protease inhibitors.R Soc Open Sci. 2025 Apr 2;12(4):241628. doi: 10.1098/rsos.241628. eCollection 2025 Apr. R Soc Open Sci. 2025. PMID: 40177100 Free PMC article.
-
An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2.Comput Struct Biotechnol J. 2021;19:4868-4883. doi: 10.1016/j.csbj.2021.08.036. Epub 2021 Aug 24. Comput Struct Biotechnol J. 2021. PMID: 34457214 Free PMC article. Review.
-
Anti-HIV reverse transcriptase plant polyphenolic natural products with in silico inhibitory properties on seven non-structural proteins vital in SARS-CoV-2 pathogenesis.J Genet Eng Biotechnol. 2021 Jul 16;19(1):104. doi: 10.1186/s43141-021-00206-2. J Genet Eng Biotechnol. 2021. PMID: 34272647 Free PMC article.
References
-
- Gorbalenya B.S., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

