Histone tails as signaling antennas of chromatin
- PMID: 33279866
- PMCID: PMC8096652
- DOI: 10.1016/j.sbi.2020.10.018
Histone tails as signaling antennas of chromatin
Abstract
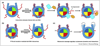
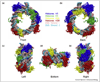
Histone tails, representing the N-terminal or C-terminal regions flanking the histone core, play essential roles in chromatin signaling networks. Intrinsic disorder of histone tails and their propensity for post-translational modifications allow them to serve as hubs in coordination of epigenetic processes within the nucleosomal context. Deposition of histone variants with distinct histone tail properties further enriches histone tails' repertoire in epigenetic signaling. Given the advances in experimental techniques and in silico modelling, we review the most recent data on histone tails' effects on nucleosome stability and dynamics, their function in regulating chromatin accessibility and folding. Finally, we discuss different molecular mechanisms to understand how histone tails are involved in nucleosome recognition by binding partners and formation of higher-order chromatin structures.
Published by Elsevier Ltd.
Figures
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389:251–260. - PubMed
-
-
Pilotto S, Speranzini V, Tortorici M, Durand D, Fish A, Valente S, Forneris F, Mai A, Sixma TK, Vachette P, et al.: Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proc Natl Acad Sci U S A 2015, 112:2752–2757.
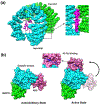
*Authors propose a tail displacement model for LSD1-mediated H3 demethylation where the interaction between the SANT2 domain and nucleosomal DNA displaces the H3 tails from DNA and make them avaibable for capture by the LSD1 active site.
-
-
-
Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, Musselman CA: The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 2018, 7.
**A study demonstrates that H3 tails extensively interact with nucleosome DNA which inhibits the binding activity of PHD finger to histone tail. It further shows that histone modifications and mutations weaken the tail-DNA interactions and increase the accessibiliy of histone tail.
-
-
- Stutzer A, Liokatis S, Kiesel A, Schwarzer D, Sprangers R, Soding J, Selenko P, Fischle W: Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol Cell 2016, 61:247–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

