Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus
- PMID: 33280122
- PMCID: PMC7986222
- DOI: 10.1111/nph.17137
Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus
Abstract
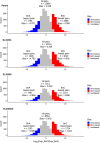
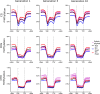
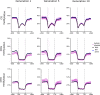
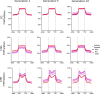
Allopolyploidisation merges evolutionarily distinct parental genomes (subgenomes) into a single nucleus. A frequent observation is that one subgenome is 'dominant' over the other subgenome, often being more highly expressed. Here, we 'replayed the evolutionary tape' with six isogenic resynthesised Brassica napus allopolyploid lines and investigated subgenome dominance patterns over the first 10 generations postpolyploidisation. We found that the same subgenome was consistently more dominantly expressed in all lines and generations and that >70% of biased gene pairs showed the same dominance patterns across all lines and an in silico hybrid of the parents. Gene network analyses indicated an enrichment for network interactions and several biological functions for the Brassica oleracea subgenome biased pairs, but no enrichment was identified for Brassica rapa subgenome biased pairs. Furthermore, DNA methylation differences between subgenomes mirrored the observed gene expression bias towards the dominant subgenome in all lines and generations. Many of these differences in gene expression and methylation were also found when comparing the progenitor genomes, suggesting that subgenome dominance is partly related to parental genome differences rather than just a byproduct of allopolyploidisation. These findings demonstrate that 'replaying the evolutionary tape' in an allopolyploid results in largely repeatable and predictable subgenome expression dominance patterns.
Keywords: Brassica napus; DNA methylation; genomic shock; hybridisation; polyploidy; rapeseed; subgenome dominance.
© 2020 The Authors New Phytologist © 2020 New Phytologist Foundation.
Figures



References
-
- Alger EI, Edger PP. 2020. One subgenome to rule them all: underlying mechanisms of subgenome dominance. Current Opinion in Plant Biology 54: 108–113. - PubMed
-
- Anderson E, Stebbins GL. 1954. Hybridization as an evolutionary stimulus. Evolution 8: 378–388.
-
- Anderson SN, Stitzer MC, Brohammer AB, Zhou P, Noshay JM, Hirsch CD, Ross‐Ibarra J, Hirsch CN, Springer NM. 2019. Transposable elements contribute to dynamic genome content in maize. The Plant Journal 100: 1052–1065. - PubMed
-
- Arnold ML, Meyer A. 2006. Natural hybridization in primates: one evolutionary mechanism. Zoology 109: 261–276. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

