Mastermind: A Comprehensive Genomic Association Search Engine for Empirical Evidence Curation and Genetic Variant Interpretation
- PMID: 33281875
- PMCID: PMC7691534
- DOI: 10.3389/fgene.2020.577152
Mastermind: A Comprehensive Genomic Association Search Engine for Empirical Evidence Curation and Genetic Variant Interpretation
Abstract
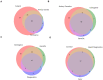
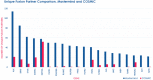
Design and interpretation of genome sequencing assays in clinical diagnostics and research labs is complicated by an inability to identify information from the medical literature and related databases quickly, comprehensively and reproducibly. This challenge is compounded by the complexity and heterogeneity of nomenclatures used to describe diseases, genes and genetic variants. Mastermind is a widely-used bioinformatic platform of genomic associations that has indexed more than 7.5 M full-text articles and 2.5 M supplemental datasets. It has automatically identified, disambiguated and annotated >6.1 M genetic variants and identified >50 K disease-gene associations. Here, we describe how Mastermind improves the sensitivity and reproducibility of clinical variant interpretation and produces comprehensive genomic landscapes of genetic variants driving pharmaceutical research. We demonstrate an alarmingly high degree of heterogeneity across commercially available panels for hereditary cancer that is resolved by evidence from Mastermind. We further examined the sensitivity of Mastermind for variant interpretation by examining 108 clinically-encountered variants and comparing the results to alternate methods. Mastermind demonstrated a sensitivity of 98.4% compared to 4.4, 45.6, and 37.4% for alternatives PubMed, Google Scholar, and ClinVar, respectively, and a specificity of 98.5% compared to 45.1, 57.6, and 68.8% as well as an increase in content yield of 22.6-, 2.2-, and 2.6-fold. When curated for clinical significance, Mastermind identified more than 4.9-fold more pathogenic variants than ClinVar for representative genes. For structural variants, we compared Mastermind's ability to sensitively identify evidence for 10 representative disease-causing CNVs versus results identified in PubMed, as well as its ability to identify evidence for fusion events compared to COSMIC. Mastermind demonstrated a 4.0- to 43.9-fold increase in references for specific CNVs compared to PubMed, as well as 5.4-fold more fusion genes when compared with COSMIC's curated database. Additionally, Mastermind produced an 8.0-fold increase in reference citations for fusion events common to Mastermind and outside databases. Taken together, these results demonstrate the utility and superiority of Mastermind in terms of both sensitivity and specificity of automated results for clinical diagnostic variant interpretation for multiple genetic variant types and highlight the potential benefit in informing pharmaceutical research.
Keywords: copy number variant; gene fusions; genome sequence analysis; oncology; rare disease; variant interpretation and classification.
Copyright © 2020 Chunn, Nefcy, Scouten, Tarpey, Chauhan, Lim, Elenitoba-Johnson, Schwartz and Kiel.
Figures
References
-
- Amendola L. M., Jarvik G. P., Leo M. C., McLaughlin H. M., Akkari Y., Amaral M. D., et al. (2016). Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am. J. Hum. Genet. 99:247. 10.1016/j.ajhg.2016.06.001 - DOI - PMC - PubMed
-
- Catalogue of Somatic Mutations in Cancer [COSMIC] (2020). Gene Fusions in Cancer. Available at: https://cancer.sanger.ac.uk/cosmic/fusion (accessed May 2020). - PMC - PubMed
-
- The Center for Applied Genomics (2020). Database of Genomic Variants - A Curated Catalogue of Human Genomic Structural Variation. Available at: http://dgv.tcag.ca/dgv/app/home (accessed July 2020).
-
- ClinGen - Clinical Genome Resource (2020). Available at: https://clinicalgenome.org/.
Grants and funding
LinkOut - more resources
Full Text Sources

