In vivo targeting of miR-223 in experimental eosinophilic oesophagitis
- PMID: 33282292
- PMCID: PMC7683276
- DOI: 10.1002/cti2.1210
In vivo targeting of miR-223 in experimental eosinophilic oesophagitis
Abstract
Objectives: Eosinophilic oesophagitis (EoE) is characterised by oesophageal inflammation, fibrosis and dysfunction. Micro (mi)-RNAs interfere with pro-inflammatory and pro-fibrotic transcriptional programs, and miR-223 was upregulated in oesophageal mucosal biopsy specimens from EoE patients. The therapeutic potential of modulating miR-223 expression in vivo has not been determined. We aimed to elucidate the relevance of oesophageal miR-223 expression in an in vivo model of EoE by inhibiting miR-223 tissue expression.
Methods: The expression of miR-223 and the validated miR-223 target insulin-like growth factor receptor 1 (IGF1R) protein was determined in our paediatric cohort of EoE patients. A murine model of Aspergillus fumigatus-induced EoE was employed, and oesophagi were assessed for miR-233, IGF1R, T lymphocyte type 2 (T2) cytokine expression and eosinophil infiltration. Mice were treated with antagomirs targeting miR-223 or resveratrol targeting its upstream regulator Midline-1(MID-1).
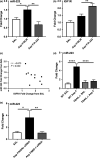
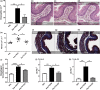
Results: There was an inverse relationship between an increased expression of miR-223 and a decreased IGF1R protein concentration in biopsy specimens from EoE patients. TNF-related apoptosis-inducing ligand deficiency, MID-1 inhibition and resveratrol treatment suppressed miR-223 expression. Furthermore, inhibition of miR-223 and treatment with resveratrol in the oesophagus resulted in an amelioration of EoE hallmark features including eosinophilic infiltration, oesophageal circumference and a reduction in T2 cytokine expression.
Conclusion: miR-223 has a key role in the perpetuation of EoE hallmark features downstream of TNF-related apoptosis-inducing ligand and MID-1 in an experimental model. These studies highlight a potentially critical role of miRNA function in EoE aetiology. miR-223 expression in the oesophagus may be therapeutically modulated by resveratrol, providing a potential new therapeutic option to be explored in EoE patients for this increasingly prevalent condition.
Keywords: Midline‐1; eosinophilic oesophagitis; microRNA; resveratrol.
© 2020 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lucendo AJ, Arias A, Gonzalez‐Cervera J et al Empiric 6‐food elimination diet induced and maintained prolonged remission in patients with adult eosinophilic esophagitis: a prospective study on the food cause of the disease. J Allergy Clin Immunol 2013; 131: 797–804. - PubMed
LinkOut - more resources
Full Text Sources
