SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus
- PMID: 33283055
- PMCID: PMC7689603
- DOI: 10.12688/wellcomeopenres.16002.2
SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus
Abstract
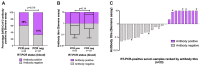
Background: Laboratory diagnosis of SARS-CoV-2 infection (the cause of COVID-19) uses PCR to detect viral RNA (vRNA) in respiratory samples. SARS-CoV-2 RNA has also been detected in other sample types, but there is limited understanding of the clinical or laboratory significance of its detection in blood. Methods: We undertook a systematic literature review to assimilate the evidence for the frequency of vRNA in blood, and to identify associated clinical characteristics. We performed RT-PCR in serum samples from a UK clinical cohort of acute and convalescent COVID-19 cases (n=212), together with convalescent plasma samples collected by NHS Blood and Transplant (NHSBT) (n=462 additional samples). To determine whether PCR-positive blood samples could pose an infection risk, we attempted virus isolation from a subset of RNA-positive samples. Results: We identified 28 relevant studies, reporting SARS-CoV-2 RNA in 0-76% of blood samples; pooled estimate 10% (95%CI 5-18%). Among serum samples from our clinical cohort, 27/212 (12.7%) had SARS-CoV-2 RNA detected by RT-PCR. RNA detection occurred in samples up to day 20 post symptom onset, and was associated with more severe disease (multivariable odds ratio 7.5). Across all samples collected ≥28 days post symptom onset, 0/494 (0%, 95%CI 0-0.7%) had vRNA detected. Among our PCR-positive samples, cycle threshold (ct) values were high (range 33.5-44.8), suggesting low vRNA copy numbers. PCR-positive sera inoculated into cell culture did not produce any cytopathic effect or yield an increase in detectable SARS-CoV-2 RNA. There was a relationship between RT-PCR negativity and the presence of total SARS-CoV-2 antibody (p=0.02). Conclusions: vRNA was detectable at low viral loads in a minority of serum samples collected in acute infection, but was not associated with infectious SARS-CoV-2 (within the limitations of the assays used). This work helps to inform biosafety precautions for handling blood products from patients with current or previous COVID-19.
Keywords: COVID-19; RNA; SARS-CoV-2; biomarker; blood; laboratory safety; viraemia; viral load.
Copyright: © 2020 Andersson MI et al.
Conflict of interest statement
Competing interests: DWE has received personal fees from Gilead, outside the submitted work.
Figures



References
-
- World Health Organisation Coronavirus disease (COVID-19) Situation Dashboard. who.int. [cited 31 Mar 2020]. Reference Source
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

