MitoEM Dataset: Large-scale 3D Mitochondria Instance Segmentation from EM Images
- PMID: 33283212
- PMCID: PMC7713709
- DOI: 10.1007/978-3-030-59722-1_7
MitoEM Dataset: Large-scale 3D Mitochondria Instance Segmentation from EM Images
Abstract
Electron microscopy (EM) allows the identification of intracellular organelles such as mitochondria, providing insights for clinical and scientific studies. However, public mitochondria segmentation datasets only contain hundreds of instances with simple shapes. It is unclear if existing methods achieving human-level accuracy on these small datasets are robust in practice. To this end, we introduce the MitoEM dataset, a 3D mitochondria instance segmentation dataset with two (30μm)3 volumes from human and rat cortices respectively, 3, 600× larger than previous benchmarks. With around 40K instances, we find a great diversity of mitochondria in terms of shape and density. For evaluation, we tailor the implementation of the average precision (AP) metric for 3D data with a 45× speedup. On MitoEM, we find existing instance segmentation methods often fail to correctly segment mitochondria with complex shapes or close contacts with other instances. Thus, our MitoEM dataset poses new challenges to the field. We release our code and data: https://donglaiw.github.io/page/mitoEM/index.html.
Keywords: 3D Instance Segmentation; EM Dataset; Mitochondria.
Figures
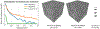
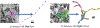

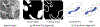

References
-
- Ariadne.ai: Automated segmentation of mitochondria and ER in cortical cells (2018. (accessed July 7, 2020)), https://ariadne.ai/case/segmentation/organelles/CorticalCells/
-
- Beier T, Pape C, Rahaman N, Prange T, Berg S, Bock DD, Cardona A, Knott GW, Plaza SM, Scheffer LK, et al.: Multicut brings automated neurite segmentation closer to human performance. Nature methods 14(2) (2017) - PubMed
-
- Chen H, Qi X, Yu L, Heng PA: DCAN: deep contour-aware networks for accurate gland segmentation In: CVPR. pp. 2487–2496. IEEE; (2016)
-
- Cheng HC, Varshney A: Volume segmentation using convolutional neural networks with limited training data In: ICIP. pp. 590–594. IEEE; (2017)
-
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3d u-net: learning dense volumetric segmentation from sparse annotation In: MICCAI. pp. 424–432. Springer; (2016)
Grants and funding
LinkOut - more resources
Full Text Sources
