Recent advances in Cannabis sativa genomics research
- PMID: 33283274
- PMCID: PMC7986631
- DOI: 10.1111/nph.17140
Recent advances in Cannabis sativa genomics research
Abstract
Cannabis (Cannabis sativa L.) is one of the oldest cultivated plants purported to have unique medicinal properties. However, scientific research of cannabis has been restricted by the Single Convention on Narcotic Drugs of 1961, an international treaty that prohibits the production and supply of narcotic drugs except under license. Legislation governing cannabis cultivation for research, medicinal and even recreational purposes has been relaxed recently in certain jurisdictions. As a result, there is now potential to accelerate cultivar development of this multi-use and potentially medically useful plant species by application of modern genomics technologies. Whilst genomics has been pivotal to our understanding of the basic biology and molecular mechanisms controlling key traits in several crop species, much work is needed for cannabis. In this review we provide a comprehensive summary of key cannabis genomics resources and their applications. We also discuss prospective applications of existing and emerging genomics technologies for accelerating the genetic improvement of cannabis.
Keywords: breeding; cannabinoids; cannabis; crop improvement; genome assembly; genomics.
© 2020 The Authors New Phytologist © 2020 New Phytologist Foundation.
Figures
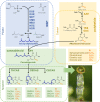
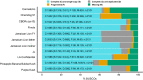

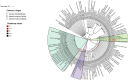
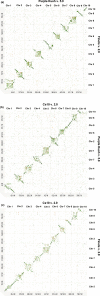

References
-
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M et al. 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology 30: 174–178. - PubMed
-
- Adams R, Hunt M, Clark J. 1940. Structure of cannabidiol, a product isolated from the marihuana extract of Minnesota wild hemp. I. Journal of the American Chemical Society 62: 196–200.
-
- Aizpurua‐Olaizola O, Soydaner U, Öztürk E, Schibano D, Simsir Y, Navarro P, Etxebarria N, Usobiaga A. 2016. Evolution of the cannabinoid and terpene content during the growth of Cannabis sativa plants from different chemotypes. Journal of Natural Products 79: 324–331. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

