3D genome evolution and reorganization in the Drosophila melanogaster species group
- PMID: 33284803
- PMCID: PMC7746282
- DOI: 10.1371/journal.pgen.1009229
3D genome evolution and reorganization in the Drosophila melanogaster species group
Abstract
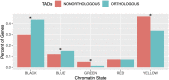
Topologically associating domains, or TADs, are functional units that organize chromosomes into 3D structures of interacting chromatin. TADs play an important role in regulating gene expression by constraining enhancer-promoter contacts and there is evidence that deletion of TAD boundaries leads to aberrant expression of neighboring genes. While the mechanisms of TAD formation have been well-studied, current knowledge on the patterns of TAD evolution across species is limited. Due to the integral role TADs play in gene regulation, their structure and organization is expected to be conserved during evolution. However, more recent research suggests that TAD structures diverge relatively rapidly. We use Hi-C chromosome conformation capture to measure evolutionary conservation of whole TADs and TAD boundary elements between D. melanogaster and D. triauraria, two early-branching species from the melanogaster species group which diverged ∼15 million years ago. We find that the majority of TADs have been reorganized since the common ancestor of D. melanogaster and D. triauraria, via a combination of chromosomal rearrangements and gain/loss of TAD boundaries. TAD reorganization between these two species is associated with a localized effect on gene expression, near the site of disruption. By separating TADs into subtypes based on their chromatin state, we find that different subtypes are evolving under different evolutionary forces. TADs enriched for broadly expressed, transcriptionally active genes are evolving rapidly, potentially due to positive selection, whereas TADs enriched for developmentally-regulated genes remain conserved, presumably due to their importance in restricting gene-regulatory element interactions. These results provide novel insight into the evolutionary dynamics of TADs and help to reconcile contradictory reports related to the evolutionary conservation of TADs and whether changes in TAD structure affect gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Lieberman-Aiden Erez, van Berkum Nynke L, Williams Louise, Imakaev Maxim, Ragoczy Tobias, Telling Agnes, Amit Ido, Lajoie Bryan R, Sabo Peter J, Dorschner Michael O, Sandstrom Richard, Bernstein Bradley, Bender M A, Groudine Mark, Gnirke Andreas, Stamatoyannopoulos John, Mirny Leonid A, Lander Eric S, and Dekker Job. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326(5950):289–293, October 2009. 10.1126/science.1181369 - DOI - PMC - PubMed
-
- Sexton Tom, Yaffe Eitan, Kenigsberg Ephraim, Bantignies Frédéric, Leblanc Benjamin, Hoichman Michael, Parrinello Hugues, Tanay Amos, and Cavalli Giacomo. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell, 148(3):458–472, February 2012. 10.1016/j.cell.2012.01.010 - DOI - PubMed
-
- Schauer Tamás, Ghavi-Helm Yad, Sexton Tom, Albig Christian, Regnard Catherine, Cavalli Giacomo, Furlong Eileen Em, and Becker Peter B. Chromosome topology guides the drosophila dosage compensation complex for target gene activation. EMBO Rep., August 2017. 10.15252/embr.201744292 - DOI - PMC - PubMed
-
- Stadler Michael R, Haines Jenna E, and Eisen Michael B. Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogaster embryo. Elife, 6, November 2017. 10.7554/eLife.29550 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

