Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles
- PMID: 33284849
- PMCID: PMC7746279
- DOI: 10.1371/journal.ppat.1009130
Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles
Abstract
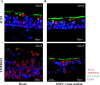
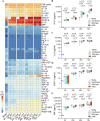
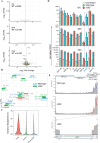
The novel coronavirus SARS-CoV-2 is the causative agent of Coronavirus Disease 2019 (COVID-19), a global healthcare and economic catastrophe. Understanding of the host immune response to SARS-CoV-2 is still in its infancy. A 382-nt deletion strain lacking ORF8 (Δ382 herein) was isolated in Singapore in March 2020. Infection with Δ382 was associated with less severe disease in patients, compared to infection with wild-type SARS-CoV-2. Here, we established Nasal Epithelial cells (NECs) differentiated from healthy nasal-tissue derived stem cells as a suitable model for the ex-vivo study of SARS-CoV-2 mediated pathogenesis. Infection of NECs with either SARS-CoV-2 or Δ382 resulted in virus particles released exclusively from the apical side, with similar replication kinetics. Screening of a panel of 49 cytokines for basolateral secretion from infected NECs identified CXCL10 as the only cytokine significantly induced upon infection, at comparable levels in both wild-type and Δ382 infected cells. Transcriptome analysis revealed the temporal up-regulation of distinct gene subsets during infection, with anti-viral signaling pathways only detected at late time-points (72 hours post-infection, hpi). This immune response to SARS-CoV-2 was significantly attenuated when compared to infection with an influenza strain, H3N2, which elicited an inflammatory response within 8 hpi, and a greater magnitude of anti-viral gene up-regulation at late time-points. Remarkably, Δ382 induced a host transcriptional response nearly identical to that of wild-type SARS-CoV-2 at every post-infection time-point examined. In accordance with previous results, Δ382 infected cells showed an absence of transcripts mapping to ORF8, and conserved expression of other SARS-CoV-2 genes. Our findings shed light on the airway epithelial response to SARS-CoV-2 infection, and demonstrate a non-essential role for ORF8 in modulating host gene expression and cytokine production from infected cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

