Robust and Scalable Learning of Complex Intrinsic Dataset Geometry via ElPiGraph
- PMID: 33286070
- PMCID: PMC7516753
- DOI: 10.3390/e22030296
Robust and Scalable Learning of Complex Intrinsic Dataset Geometry via ElPiGraph
Abstract
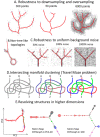
Multidimensional datapoint clouds representing large datasets are frequently characterized by non-trivial low-dimensional geometry and topology which can be recovered by unsupervised machine learning approaches, in particular, by principal graphs. Principal graphs approximate the multivariate data by a graph injected into the data space with some constraints imposed on the node mapping. Here we present ElPiGraph, a scalable and robust method for constructing principal graphs. ElPiGraph exploits and further develops the concept of elastic energy, the topological graph grammar approach, and a gradient descent-like optimization of the graph topology. The method is able to withstand high levels of noise and is capable of approximating data point clouds via principal graph ensembles. This strategy can be used to estimate the statistical significance of complex data features and to summarize them into a single consensus principal graph. ElPiGraph deals efficiently with large datasets in various fields such as biology, where it can be used for example with single-cell transcriptomic or epigenomic datasets to infer gene expression dynamics and recover differentiation landscapes.
Keywords: data approximation; principal graphs; principal trees; software; topological grammars.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Roux B.L., Rouanet H. Geometric Data Analysis: From Correspondence Analysis to Structured Data Analysis. Springer; Dordrecht, The Netherlands: 2005.
-
- Gorban A., Kégl B., Wunch D., Zinovyev A. Principal Manifolds for Data Visualisation and Dimension Reduction. Springer; Berlin/Heidelberg, Germany: 2008.
-
- Carlsson G. Topology and data. Bull. Am. Math. Soc. 2009;46:255–308. doi: 10.1090/S0273-0979-09-01249-X. - DOI
-
- Camastra F., Staiano A. Intrinsic dimension estimation: Advances and open problems. Inf. Sci. 2016;328:26–41. doi: 10.1016/j.ins.2015.08.029. - DOI
LinkOut - more resources
Full Text Sources

