Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks
- PMID: 33286504
- PMCID: PMC7517272
- DOI: 10.3390/e22070732
Information Transfer in Linear Multivariate Processes Assessed through Penalized Regression Techniques: Validation and Application to Physiological Networks
Abstract
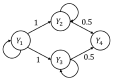
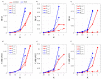
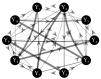
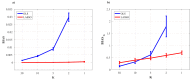
The framework of information dynamics allows the dissection of the information processed in a network of multiple interacting dynamical systems into meaningful elements of computation that quantify the information generated in a target system, stored in it, transferred to it from one or more source systems, and modified in a synergistic or redundant way. The concepts of information transfer and modification have been recently formulated in the context of linear parametric modeling of vector stochastic processes, linking them to the notion of Granger causality and providing efficient tools for their computation based on the state-space (SS) representation of vector autoregressive (VAR) models. Despite their high computational reliability these tools still suffer from estimation problems which emerge, in the case of low ratio between data points available and the number of time series, when VAR identification is performed via the standard ordinary least squares (OLS). In this work we propose to replace the OLS with penalized regression performed through the Least Absolute Shrinkage and Selection Operator (LASSO), prior to computation of the measures of information transfer and information modification. First, simulating networks of several coupled Gaussian systems with complex interactions, we show that the LASSO regression allows, also in conditions of data paucity, to accurately reconstruct both the underlying network topology and the expected patterns of information transfer. Then we apply the proposed VAR-SS-LASSO approach to a challenging application context, i.e., the study of the physiological network of brain and peripheral interactions probed in humans under different conditions of rest and mental stress. Our results, which document the possibility to extract physiologically plausible patterns of interaction between the cardiovascular, respiratory and brain wave amplitudes, open the way to the use of our new analysis tools to explore the emerging field of Network Physiology in several practical applications.
Keywords: State–space models; conditional transfer entropy; entropy; information dynamics; linear prediction; multivariate time series analysis; network physiology; partial information decomposition; penalized regression techniques; vector autoregressive model.
Conflict of interest statement
The authors declare no conflict of interests.
Figures

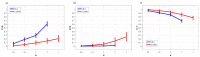


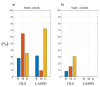
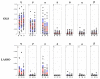
References
-
- Faes L., Porta A., Nollo G. Information decomposition in bivariate systems: Theory and application to cardiorespiratory dynamics. Entropy. 2015;17:277–303. doi: 10.3390/e17010277. - DOI
-
- Malik M. Heart rate variability: Standards of measurement, physiological interpretation, and clinical use: Task force of the European Society of Cardiology and the North American Society for Pacing and Electrophysiology. Ann. Noninvasive Electrocardiol. 1996;1:151–181. doi: 10.1111/j.1542-474X.1996.tb00275.x. - DOI
LinkOut - more resources
Full Text Sources

