Standardization of an LNA-based TaqMan assay qPCR analysis for Aspiculuris tetraptera DNA in mouse faeces
- PMID: 33287731
- PMCID: PMC7720592
- DOI: 10.1186/s12866-020-02053-6
Standardization of an LNA-based TaqMan assay qPCR analysis for Aspiculuris tetraptera DNA in mouse faeces
Abstract
Background: Aspiculuris tetraptera, as a parasitic pinworm, is most frequently detected in laboratory mice, and transmission is mediated by the eggs contained in the faeces of infected mice. A highly sensitive and quantitative faeces-based diagnostic tool would be useful for the early detection of A. tetraptera to inhibit the expansion of infection. In this study, we developed a quantitative assay that exhibits high sensitivity in detecting A. tetraptera in faeces using PCR techniques.
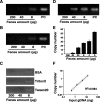
Results: Endpoint PCR demonstrated the detection of A. tetraptera DNA in 0.5 ng genomic DNA extracted from the faeces of infected mice. To quantitatively detect the small amount of A. tetraptera DNA, locked nucleic acid (LNA)-based primers and LNA-based TaqMan probes were used for the quantitative PCR assay (qPCR). The combination of LNA-based DNA increased detection sensitivity by more than 100-fold compared to using normal oligo DNAs. The copy number of the A. tetraptera DNA detected was positively related to the infected faeces-derived genomic DNA with a simple linearity regression in the range of 20 pg to 15 ng of the genomic DNA. To more conveniently detect infection using faeces, the LNA-based TaqMan assay was applied to the crude fraction of the faeces without DNA purification. An assay using ethanol precipitation of the faeces yielded results consistent with those of direct microscopic observation.
Conclusion: The LNA-TaqMan assay developed in this study quantitatively detects A. tetraptera infection in mouse faeces.
Keywords: A. tetraptera; Faeces; Locked nucleic acid; Mice; PCR; TaqMan assay.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 17K17918/Research for Promoting Technological Seeds of Japan Science and Technology Corporation (JST) Ministry of Education
- 18K16255/Research for Promoting Technological Seeds of Japan Science and Technology Corporation (JST) Ministry of Education
- 18K11128/Research for Promoting Technological Seeds of Japan Science and Technology Corporation (JST) Ministry of Education
LinkOut - more resources
Full Text Sources
Medical

