ARP2/3-independent WAVE/SCAR pathway and class XI myosin control sperm nuclear migration in flowering plants
- PMID: 33288691
- PMCID: PMC7768783
- DOI: 10.1073/pnas.2015550117
ARP2/3-independent WAVE/SCAR pathway and class XI myosin control sperm nuclear migration in flowering plants
Abstract
After eukaryotic fertilization, gamete nuclei migrate to fuse parental genomes in order to initiate development of the next generation. In most animals, microtubules control female and male pronuclear migration in the zygote. Flowering plants, on the other hand, have evolved actin filament (F-actin)-based sperm nuclear migration systems for karyogamy. Flowering plants have also evolved a unique double-fertilization process: two female gametophytic cells, the egg and central cells, are each fertilized by a sperm cell. The molecular and cellular mechanisms of how flowering plants utilize and control F-actin for double-fertilization events are largely unknown. Using confocal microscopy live-cell imaging with a combination of pharmacological and genetic approaches, we identified factors involved in F-actin dynamics and sperm nuclear migration in Arabidopsis thaliana (Arabidopsis) and Nicotiana tabacum (tobacco). We demonstrate that the F-actin regulator, SCAR2, but not the ARP2/3 protein complex, controls the coordinated active F-actin movement. These results imply that an ARP2/3-independent WAVE/SCAR-signaling pathway regulates F-actin dynamics in female gametophytic cells for fertilization. We also identify that the class XI myosin XI-G controls active F-actin movement in the Arabidopsis central cell. XI-G is not a simple transporter, moving cargos along F-actin, but can generate forces that control the dynamic movement of F-actin for fertilization. Our results provide insights into the mechanisms that control gamete nuclear migration and reveal regulatory pathways for dynamic F-actin movement in flowering plants.
Keywords: F-actin; WAVE/SCAR; fertilization; myosin; nuclear migration.
Conflict of interest statement
The authors declare no competing interest.
Figures


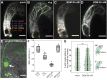
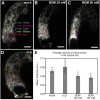
References
-
- Reinsch S., Gönczy P., Mechanisms of nuclear positioning. J. Cell Sci. 111, 2283–2295 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

