Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing
- PMID: 33288908
- PMCID: PMC7610403
- DOI: 10.1038/s41593-020-00736-x
Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing
Abstract
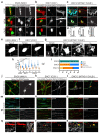
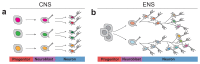
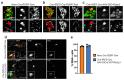
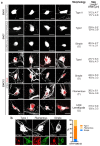
Autonomous regulation of the intestine requires the combined activity of functionally distinct neurons of the enteric nervous system (ENS). However, the variety of enteric neuron types and how they emerge during development remain largely unknown. Here, we define a molecular taxonomy of 12 enteric neuron classes within the myenteric plexus of the mouse small intestine using single-cell RNA sequencing. We present cell-cell communication features and histochemical markers for motor neurons, sensory neurons and interneurons, together with transgenic tools for class-specific targeting. Transcriptome analysis of the embryonic ENS uncovers a novel principle of neuronal diversification, where two neuron classes arise through a binary neurogenic branching and all other identities emerge through subsequent postmitotic differentiation. We identify generic and class-specific transcriptional regulators and functionally connect Pbx3 to a postmitotic fate transition. Our results offer a conceptual and molecular resource for dissecting ENS circuits and predicting key regulators for directed differentiation of distinct enteric neuron classes.
Conflict of interest statement
The authors disclose no conflicts.
Figures













Comment in
-
Revealing the complexity of the gut's brain.Nat Neurosci. 2021 Jan;24(1):1-2. doi: 10.1038/s41593-020-00769-2. Nat Neurosci. 2021. PMID: 33303973 No abstract available.
References
-
- Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9:286–294. - PubMed
-
- Veiga-Fernandes H, Pachnis V. Neuroimmune regulation during intestinal development and homeostasis. Nature Immunology. 2017;18:116–122. - PubMed
-
- Furness JB. Types of neurons in the enteric nervous system. J Auton Nerv Syst. 2000;81:87–96. - PubMed
-
- Sang Q, Young HM. Chemical coding of neurons in the myenteric plexus and external muscle of the small and large intestine of the mouse. Cell Tissue Res. 1996;284:39–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

