A small molecule that mitigates bacterial infection disrupts Gram-negative cell membranes and is inhibited by cholesterol and neutral lipids
- PMID: 33290418
- PMCID: PMC7748285
- DOI: 10.1371/journal.ppat.1009119
A small molecule that mitigates bacterial infection disrupts Gram-negative cell membranes and is inhibited by cholesterol and neutral lipids
Abstract
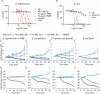
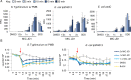
Infections caused by Gram-negative bacteria are difficult to fight because these pathogens exclude or expel many clinical antibiotics and host defense molecules. However, mammals have evolved a substantial immune arsenal that weakens pathogen defenses, suggesting the feasibility of developing therapies that work in concert with innate immunity to kill Gram-negative bacteria. Using chemical genetics, we recently identified a small molecule, JD1, that kills Salmonella enterica serovar Typhimurium (S. Typhimurium) residing within macrophages. JD1 is not antibacterial in standard microbiological media, but rapidly inhibits growth and curtails bacterial survival under broth conditions that compromise the outer membrane or reduce efflux pump activity. Using a combination of cellular indicators and super resolution microscopy, we found that JD1 damaged bacterial cytoplasmic membranes by increasing fluidity, disrupting barrier function, and causing the formation of membrane distortions. We quantified macrophage cell membrane integrity and mitochondrial membrane potential and found that disruption of eukaryotic cell membranes required approximately 30-fold more JD1 than was needed to kill bacteria in macrophages. Moreover, JD1 preferentially damaged liposomes with compositions similar to E. coli inner membranes versus mammalian cell membranes. Cholesterol, a component of mammalian cell membranes, was protective in the presence of neutral lipids. In mice, intraperitoneal administration of JD1 reduced tissue colonization by S. Typhimurium. These observations indicate that during infection, JD1 gains access to and disrupts the cytoplasmic membrane of Gram-negative bacteria, and that neutral lipids and cholesterol protect mammalian membranes from JD1-mediated damage. Thus, it may be possible to develop therapeutics that exploit host innate immunity to gain access to Gram-negative bacteria and then preferentially damage the bacterial cell membrane over host membranes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

