Regional, Layer, and Cell-Type-Specific Connectivity of the Mouse Default Mode Network
- PMID: 33290731
- PMCID: PMC8150331
- DOI: 10.1016/j.neuron.2020.11.011
Regional, Layer, and Cell-Type-Specific Connectivity of the Mouse Default Mode Network
Abstract
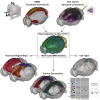
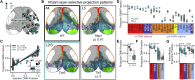
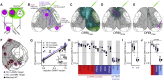
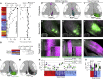
The evolutionarily conserved default mode network (DMN) is a distributed set of brain regions coactivated during resting states that is vulnerable to brain disorders. How disease affects the DMN is unknown, but detailed anatomical descriptions could provide clues. Mice offer an opportunity to investigate structural connectivity of the DMN across spatial scales with cell-type resolution. We co-registered maps from functional magnetic resonance imaging and axonal tracing experiments into the 3D Allen mouse brain reference atlas. We find that the mouse DMN consists of preferentially interconnected cortical regions. As a population, DMN layer 2/3 (L2/3) neurons project almost exclusively to other DMN regions, whereas L5 neurons project in and out of the DMN. In the retrosplenial cortex, a core DMN region, we identify two L5 projection types differentiated by in- or out-DMN targets, laminar position, and gene expression. These results provide a multi-scale description of the anatomical correlates of the mouse DMN.
Keywords: DMN; Default mode network; axonal projections; connectivity; cortical connectome; projection neuron types; retrosplenial cortex; single cell transcriptomics; viral tracer.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

