Endurance Runners with Intramyocellular Lipid Accumulation and High Insulin Sensitivity Have Enhanced Expression of Genes Related to Lipid Metabolism in Muscle
- PMID: 33291227
- PMCID: PMC7762159
- DOI: 10.3390/jcm9123951
Endurance Runners with Intramyocellular Lipid Accumulation and High Insulin Sensitivity Have Enhanced Expression of Genes Related to Lipid Metabolism in Muscle
Abstract
Context: Endurance-trained athletes have high oxidative capacities, enhanced insulin sensitivities, and high intracellular lipid accumulation in muscle. These characteristics are likely due to altered gene expression levels in muscle.
Design and setting: We compared intramyocellular lipid (IMCL), insulin sensitivity, and gene expression levels of the muscle in eight nonobese healthy men (control group) and seven male endurance athletes (athlete group). Their IMCL levels were measured by proton-magnetic resonance spectroscopy, and their insulin sensitivity was evaluated by glucose infusion rate (GIR) during a euglycemic-hyperinsulinemic clamp. Gene expression levels in the vastus lateralis were evaluated by quantitative RT-PCR (qRT-PCR) and microarray analysis.
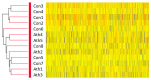
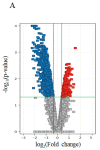
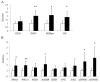
Results: IMCL levels in the tibialis anterior muscle were approximately 2.5 times higher in the athlete group compared to the control group, while the IMCL levels in the soleus muscle and GIR were comparable. In the microarray hierarchical clustering analysis, gene expression patterns were not clearly divided into control and athlete groups. In a gene set enrichment analysis with Gene Ontology gene sets, "RESPONSE TO LIPID" was significantly upregulated in the athlete group compared with the control group. Indeed, qRT-PCR analysis revealed that, compared to the control group, the athlete group had 2-3 times higher expressions of proliferator-activated receptor gamma coactivator-1 alpha (PGC1A), adiponectin receptors (AdipoRs), and fatty acid transporters including fatty acid transporter-1, plasma membrane-associated fatty acid binding protein, and lipoprotein lipase.
Conclusions: Endurance runners with higher IMCL levels have higher expression levels of genes related to lipid metabolism such as PGC1A, AdipoRs, and fatty acid transporters in muscle.
Keywords: adiponectin receptor; athlete’s paradox; insulin resistance; intramyocellular lipid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Association between expression of FABPpm in skeletal muscle and insulin sensitivity in intramyocellular lipid-accumulated nonobese men.J Clin Endocrinol Metab. 2014 Sep;99(9):3343-52. doi: 10.1210/jc.2014-1896. Epub 2014 Jun 17. J Clin Endocrinol Metab. 2014. PMID: 24937540
-
Increased intramyocellular lipid/impaired insulin sensitivity is associated with altered lipid metabolic genes in muscle of high responders to a high-fat diet.Am J Physiol Endocrinol Metab. 2016 Jan 1;310(1):E32-40. doi: 10.1152/ajpendo.00220.2015. Epub 2015 Oct 20. Am J Physiol Endocrinol Metab. 2016. PMID: 26487001 Clinical Trial.
-
Dissociation of intramyocellular lipid storage and insulin resistance in trained athletes and type 2 diabetes patients; involvement of perilipin 5?J Physiol. 2018 Mar 1;596(5):857-868. doi: 10.1113/JP275182. Epub 2017 Nov 23. J Physiol. 2018. PMID: 29110300 Free PMC article.
-
Both higher fitness level and higher current physical activity level may be required for intramyocellular lipid accumulation in non-athlete men.Sci Rep. 2020 Mar 5;10(1):4102. doi: 10.1038/s41598-020-61080-5. Sci Rep. 2020. PMID: 32139784 Free PMC article.
-
Intramyocellular lipid content in human skeletal muscle.Obesity (Silver Spring). 2006 Mar;14(3):357-67. doi: 10.1038/oby.2006.47. Obesity (Silver Spring). 2006. PMID: 16648604 Review.
Cited by
-
Role of the PPARGC1A Gene and Its rs8192678 Polymorphism on Sport Performance, Aerobic Capacity, Muscle Adaptation and Metabolic Diseases: A Narrative Review.Genes (Basel). 2024 Dec 20;15(12):1631. doi: 10.3390/genes15121631. Genes (Basel). 2024. PMID: 39766897 Free PMC article. Review.
-
Regulation of PXR Function by Coactivator and Corepressor Proteins: Ligand Binding Is Just the Beginning.Cells. 2021 Nov 12;10(11):3137. doi: 10.3390/cells10113137. Cells. 2021. PMID: 34831358 Free PMC article. Review.
-
Optimised Skeletal Muscle Mass as a Key Strategy for Obesity Management.Metabolites. 2025 Feb 1;15(2):85. doi: 10.3390/metabo15020085. Metabolites. 2025. PMID: 39997710 Free PMC article. Review.
References
-
- Kakehi S., Tamura Y., Takeno K., Sakurai Y., Kawaguchi M., Watanabe T., Funayama T., Sato F., Ikeda S., Kanazawa A., et al. Increased intramyocellular lipid/impaired insulin sensitivity is associated with altered lipid metabolic genes in muscle of high responders to a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2016;310:32–40. doi: 10.1152/ajpendo.00220.2015. - DOI - PubMed
-
- O’Leary V.B., Jorett A.E., Marchetti C.M., Gonzalez F., Phillips S.A., Ciaraldi T.P., Kirwan J.P. Enhanced adiponectin multimer ratio and skeletal muscle adiponectin receptor expression following exercise training and diet in older insulin-resistant adults. Am. J. Physiol. 2007;293:421–427. doi: 10.1152/ajpendo.00123.2007. - DOI - PubMed
-
- Christiansen T., Paulsen S.K., Bruun J.M., Ploug T., Pedersen S.B., Richelsen B. Diet-induced weight loss and exercise alone and in combination enhance the expression of adiponectin receptors in adipose tissue and skeletal muscle, but only diet-induced weight loss enhanced circulating adiponectin. J. Clin. Endocrinol. Metab. 2010;95:911–919. doi: 10.1210/jc.2008-2505. - DOI - PubMed
-
- Bluher M., Bullen J.W., Jr., Lee J.H., Kralisch S., Fasshauer M., Kloting N., Niebauer J., Schon M.R., Williams C.J., Mantzoros C.S. Circulating adiponectin and expression of adiponectin receptors in human skeletal muscle: Associations with metabolic parameters and insulin resistance and regulation by physical training. J. Clin. Endocrinol. Metab. 2006;91:2310–2316. doi: 10.1210/jc.2005-2556. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

