The T3SS of Shigella: Expression, Structure, Function, and Role in Vacuole Escape
- PMID: 33291504
- PMCID: PMC7762205
- DOI: 10.3390/microorganisms8121933
The T3SS of Shigella: Expression, Structure, Function, and Role in Vacuole Escape
Abstract
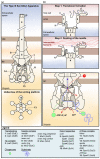
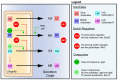
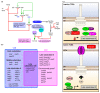
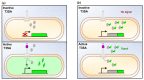
Shigella spp. are one of the leading causes of infectious diarrheal diseases. They are Escherichia coli pathovars that are characterized by the harboring of a large plasmid that encodes most virulence genes, including a type III secretion system (T3SS). The archetypal element of the T3SS is the injectisome, a syringe-like nanomachine composed of approximately 20 proteins, spanning both bacterial membranes and the cell wall, and topped with a needle. Upon contact of the tip of the needle with the plasma membrane, the injectisome secretes its protein substrates into host cells. Some of these substrates act as translocators or effectors whose functions are key to the invasion of the cytosol and the cell-to-cell spread characterizing the lifestyle of Shigella spp. Here, we review the structure, assembly, function, and methods to measure the activity of the injectisome with a focus on Shigella, but complemented with data from other T3SS if required. We also present the regulatory cascade that controls the expression of T3SS genes in Shigella. Finally, we describe the function of translocators and effectors during cell-to-cell spread, particularly during escape from the vacuole, a key element of Shigella's pathogenesis that has yet to reveal all of its secrets.
Keywords: Shigella; autophagy; genetically encoded reporter; injectisome; secretion; transcription regulation; type III secretion system (T3SS); vacuole rupture; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

