Circ0120816 acts as an oncogene of esophageal squamous cell carcinoma by inhibiting miR-1305 and releasing TXNRD1
- PMID: 33292234
- PMCID: PMC7597039
- DOI: 10.1186/s12935-020-01617-w
Circ0120816 acts as an oncogene of esophageal squamous cell carcinoma by inhibiting miR-1305 and releasing TXNRD1
Abstract
Background: Circular RNAs (circRNAs) have been discovered to participate in the carcinogenesis of multiple cancers. However, the role of circRNAs in esophageal squamous cell carcinoma (ESCC) progression is yet to be properly understood. This research aimed to investigate and understand the mechanism used by circRNAs to regulate ESCC progression.
Methods: Bioinformatics analysis was first performed to screen dysregulated circRNAs and differentially expressed genes in ESCC. The ESCC tissue samples and adjacent normal tissue samples utilized in this study were obtained from 36 ESCC patients. All the samples were subjected to qRT-PCR analysis to identify the expression of TXNRD1, circRNAs, and miR-1305. Luciferase reporter assay, RNA immunoprecipitation assay and RNA pull-down assay were later conducted to verify the existing relationship among circ0120816, miR-1305 and TXNRD1. CCK-8, BrdU, cell adhesion, cell cycle, western blot and caspase 3 activity assays were also employed to evaluate the regulation of these three biological molecules in ESCC carcinogenesis. To evaluate the effect of circ0120816 on ESCC tumor growth and metastasis, the xenograft mice model was constructed.
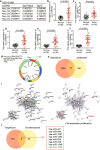
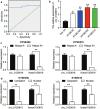
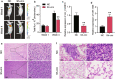
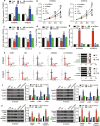
Results: Experimental investigations revealed that circ0120816 was the highest upregulated circRNA in ESCC tissues and that this non-coding RNA acted as a miR-1305 sponge in enhancing cell viability, cell proliferation, and cell adhesion as well as repressing cell apoptosis in ESCC cell lines. Moreover, miR-1305 was observed to exert a tumor-suppressive effect in ESCC cells by directly targeting and repressing TXNRD1. It was also noticed that TXNRD1 could regulate cyclin, cell adhesion molecule, and apoptosis-related proteins. Furthermore, silencing circ0120816 was found to repress ESCC tumor growth and metastasis in vivo.
Conclusions: This research confirmed that circ0120816 played an active role in promoting ESCC development by targeting miR-1305 and upregulating oncogene TXNRD1.
Keywords: Esophageal squamous cell carcinoma; TXNRD1; circ0120816; miR-1305.
Conflict of interest statement
The authors declare that no conflict of interests exist in this research project.
Figures



Similar articles
-
Exosomal circRNA FNDC3B promotes the progression of esophageal squamous cell carcinoma by sponging miR-490-5p and regulating thioredoxin reductase 1 expression.Bioengineered. 2022 May;13(5):13829-13848. doi: 10.1080/21655979.2022.2084484. Bioengineered. 2022. PMID: 35703190 Free PMC article.
-
CircRNA_2646 functions as a ceRNA to promote progression of esophageal squamous cell carcinoma via inhibiting miR-124/PLP2 signaling pathway.Cell Death Discov. 2021 May 11;7(1):99. doi: 10.1038/s41420-021-00461-9. Cell Death Discov. 2021. PMID: 33976115 Free PMC article.
-
Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma.J Exp Clin Cancer Res. 2017 Nov 16;36(1):161. doi: 10.1186/s13046-017-0622-1. J Exp Clin Cancer Res. 2017. PMID: 29145896 Free PMC article.
-
Current advances and future perspectives on the functional roles and clinical implications of circular RNAs in esophageal squamous cell carcinoma: more influential than expected.Biomark Res. 2022 Jun 7;10(1):41. doi: 10.1186/s40364-022-00388-y. Biomark Res. 2022. PMID: 35672804 Free PMC article. Review.
-
Roles of circRNA dysregulation in esophageal squamous cell carcinoma tumor microenvironment.Front Oncol. 2023 Jun 13;13:1153207. doi: 10.3389/fonc.2023.1153207. eCollection 2023. Front Oncol. 2023. PMID: 37384299 Free PMC article. Review.
Cited by
-
CircSFMBT2 Plays an Oncogenic Role in Lung Adenocarcinoma Depending on the miR-1305/SALL4 Axis.Biochem Genet. 2024 Oct;62(5):3485-3503. doi: 10.1007/s10528-023-10611-6. Epub 2023 Dec 21. Biochem Genet. 2024. PMID: 38127171
-
Circular RNAs in Cell Cycle Regulation of Cancers.Int J Mol Sci. 2024 May 31;25(11):6094. doi: 10.3390/ijms25116094. Int J Mol Sci. 2024. PMID: 38892280 Free PMC article. Review.
-
Hsa_circ_0072008 regulates cell proliferation, migration, and invasion in cervical squamous cell carcinoma via miR-1305/helicase, lymphoid specific (HELLS) axis.Bioengineered. 2022 Apr;13(4):8311-8322. doi: 10.1080/21655979.2022.2048945. Bioengineered. 2022. PMID: 35311456 Free PMC article.
-
Resveratrol Contrasts IL-6 Pro-Growth Effects and Promotes Autophagy-Mediated Cancer Cell Dormancy in 3D Ovarian Cancer: Role of miR-1305 and of Its Target ARH-I.Cancers (Basel). 2022 Apr 25;14(9):2142. doi: 10.3390/cancers14092142. Cancers (Basel). 2022. PMID: 35565270 Free PMC article.
-
Regulation of ferroptosis by noncoding RNAs: a novel promise treatment in esophageal squamous cell carcinoma.Mol Cell Biochem. 2022 Sep;477(9):2193-2202. doi: 10.1007/s11010-022-04441-0. Epub 2022 Apr 21. Mol Cell Biochem. 2022. PMID: 35449482 Review.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

