LigGrep: a tool for filtering docked poses to improve virtual-screening hit rates
- PMID: 33292486
- PMCID: PMC7656723
- DOI: 10.1186/s13321-020-00471-2
LigGrep: a tool for filtering docked poses to improve virtual-screening hit rates
Abstract
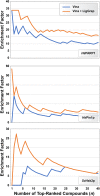
Structure-based virtual screening (VS) uses computer docking to prioritize candidate small-molecule ligands for subsequent experimental testing. Docking programs evaluate molecular binding in part by predicting the geometry with which a given compound might bind a target receptor (e.g., the docked "pose" relative to a protein target). Candidate ligands predicted to participate in the same intermolecular interactions typical of known ligands (or ligands that bind related proteins) are arguably more likely to be true binders. Some docking programs allow users to apply constraints during the docking process with the goal of prioritizing these critical interactions. But these programs often have restrictive and/or expensive licenses, and many popular open-source docking programs (e.g., AutoDock Vina) lack this important functionality. We present LigGrep, a free, open-source program that addresses this limitation. As input, LigGrep accepts a protein receptor file, a directory containing many docked-compound files, and a list of user-specified filters describing critical receptor/ligand interactions. LigGrep evaluates each docked pose and outputs the names of the compounds with poses that pass all filters. To demonstrate utility, we show that LigGrep can improve the hit rates of test VS targeting H. sapiens poly(ADPribose) polymerase 1 (HsPARP1), H. sapiens peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (HsPin1p), and S. cerevisiae hexokinase-2 (ScHxk2p). We hope that LigGrep will be a useful tool for the computational biology community. A copy is available free of charge at http://durrantlab.com/liggrep/ .
Keywords: Computational biology; Computer-aided drug discovery; Filters; Virtual screening.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Kontoyianni M. Docking and virtual screening in drug discovery. Methods Mol Biol. 2017;1647:255–266. - PubMed
-
- Varghese JN, Smith PW, Sollis SL, Blick TJ, Sahasrabudhe A, McKimm-Breschkin JL, Colman PM. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure. 1998;6:735–46. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

